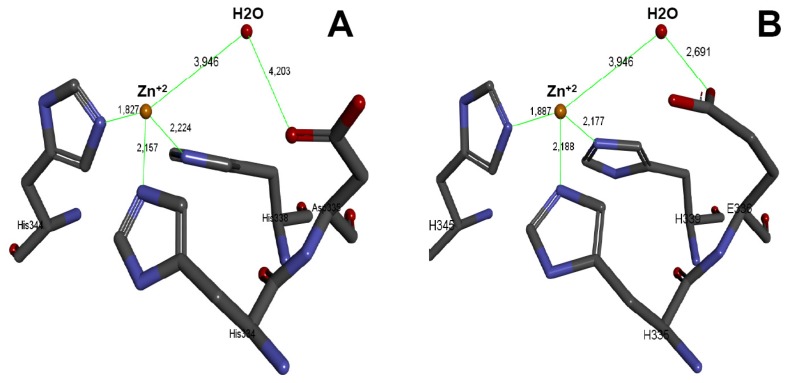
Figure 6.
3D model of the active site HDLGHNLCIDH. Using the Discovery Studio Program Version 3.5.0.12158 (Accelrys Software Inc, San Diego, CA, USA), a 3D model of the active site of BlatPII was generated (A). Determination of distances in the predicted model was made using the coordinates of the zinc atom and the water molecule from the crystallographic structure of VAP-1 from venom of Crotalus atrox (B).