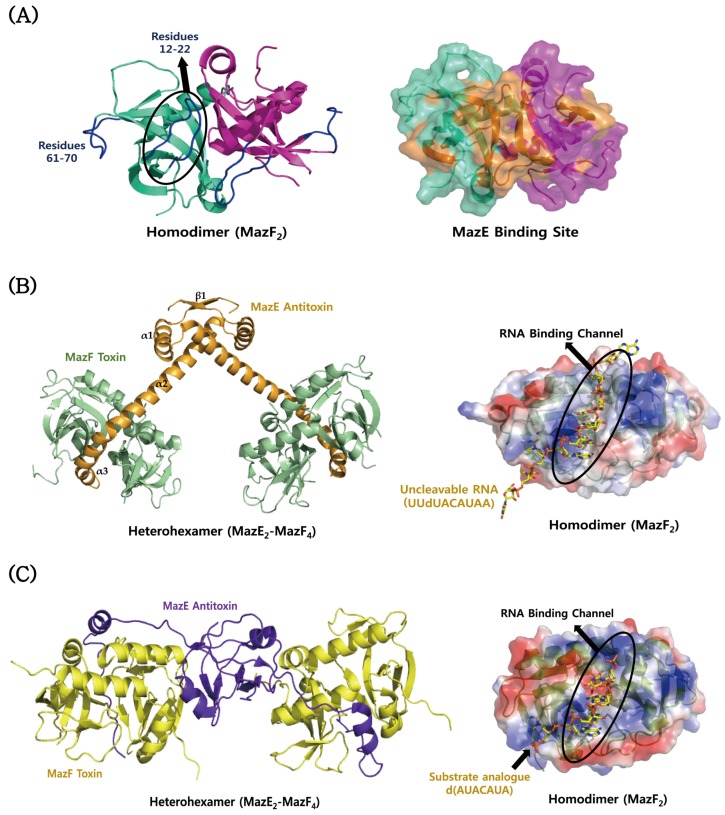
Figure 2.
Structural comparisons of MazEF proteins from S. aureus, B. subtilis, and E. coli. (A) Structure of S. aureus MazF. The residues showing a relatively high mobility are indicated in blue. MazE binding site is mapped in orange onto the surface model of MazF. (B) Structure of B. subtilis MazF in complex with B. subtilis MazE (left panel) and its uncleavable RNA substrate (UUdUACAUAA) (right panel); (C) structure of E. coli MazF in complex with E. coli MazE (left panel) and the substrate analogue [d(AUACAUA)] (right panel). In (B) and (C), an RNA binding channel of MazF is formed by its dimerization.