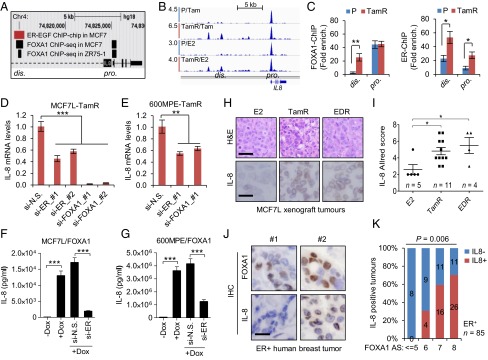
Fig. 6.
Increased FOXA1 and ER regulate IL-8 expression in ER+ breast cancer. (A) Schematic diagram of ER and FOXA1 binding within the IL8 gene locus as defined by EGF-stimulated ER ChIP-on-chip (37) and FOXA1 ChIP-sEq. (7) in MCF7 cells. (B) Snapshot of FOXA1 continuous peaks from ChIP-seq data showing the binding pattern upstream of the IL8 gene TSS in MCF7L-P and MCF7L-TamR cells treated with Tam or E2. (C) FOXA1-ChIP (Left) and ER-ChIP (Right) followed by qPCR of binding regions in MCF7L-P and TamR cells. Quantification of amplified binding regions was calculated as fold enrichment by normalizing to an intergenic sequence as a negative control. (D and E) Measurement of IL-8 mRNA by qRT-PCR in MCF7L-TamR and 600MPE-TamR cells with either ER or FOXA1 knockdown. N.S., nonspecific; #1 and #2, two different siRNA sequences. (F and G) ELISA of IL-8 protein in culture media of MCF7L/FOXA1 and 600MPE/FOXA1 cells −/+Dox in the absence/presence of ER knockdown. (H) Representative H&E staining and IL-8 IHC images from E2-treated and Endo-R MCF7L xenograft tumors. (Scale bars, 100 μm and 50 μm, respectively.) (I) Scatter dot plots of IL-8 Allred score in H. Data represent means ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, two-sided t test for indicated comparisons. (J) Representative IHC images from two ER+ tumors showing low (#1) vs. high (#2) FOXA1 and the negative vs. positive IL-8 staining, respectively. (Scale bar, 50 μm.) (K) Proportions of positive vs. negative IL-8 tumors within the groups of tumors showing the same FOXA1 Allred score (AS). Correlation of IL-8 positivity and FOXA1-AS was evaluated by Fisher’s exact test.