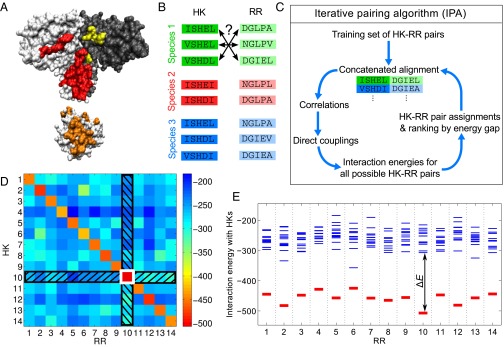
Fig. 1.
Iterative pairing algorithm (IPA). (A) Surface representations of an HK dimer (top) and an RR (bottom), from a cocrystal structure (27); the HK−RR contacts in each molecule are highlighted in color. (B) To correctly pair HKs and RRs in each species from their sequences alone, we start from multiple sequence alignments of HKs and RRs, including 64 amino acids from the HK and 112 from the RR. (C) Schematic of the main steps of the IPA. (D and E) Example of HK−RR pair assignment and ranking by energy gap for one species. (D) Color map of the matrix of HK−RR interaction energies in E. coli K-12 MG1655 from the final iteration of the IPA performed on our standard dataset, with a training set of HK−RR pairs, and an increment step of pairs. As in every IPA iteration and every species, the pair with the lowest interaction energy is selected first (here, HK 10 and RR 10, boxed in white), and this HK and RR are removed from further consideration (black hatches). Then, the next pair with the lowest energy is chosen, and the process is repeated until all HKs and RRs are paired. (E) Energy spectrum from D, showing the interaction energies with all of the HKs for each RR, with the correct HK−RR pairs shown in red. The energy gap is shown for RR 10. A confidence score based on the energy gap is used to rank all assigned HK−RR pairs, and this ranking is exploited to build the CA for the subsequent IPA iteration. See Materials and Methods for details.