Fig. 1.
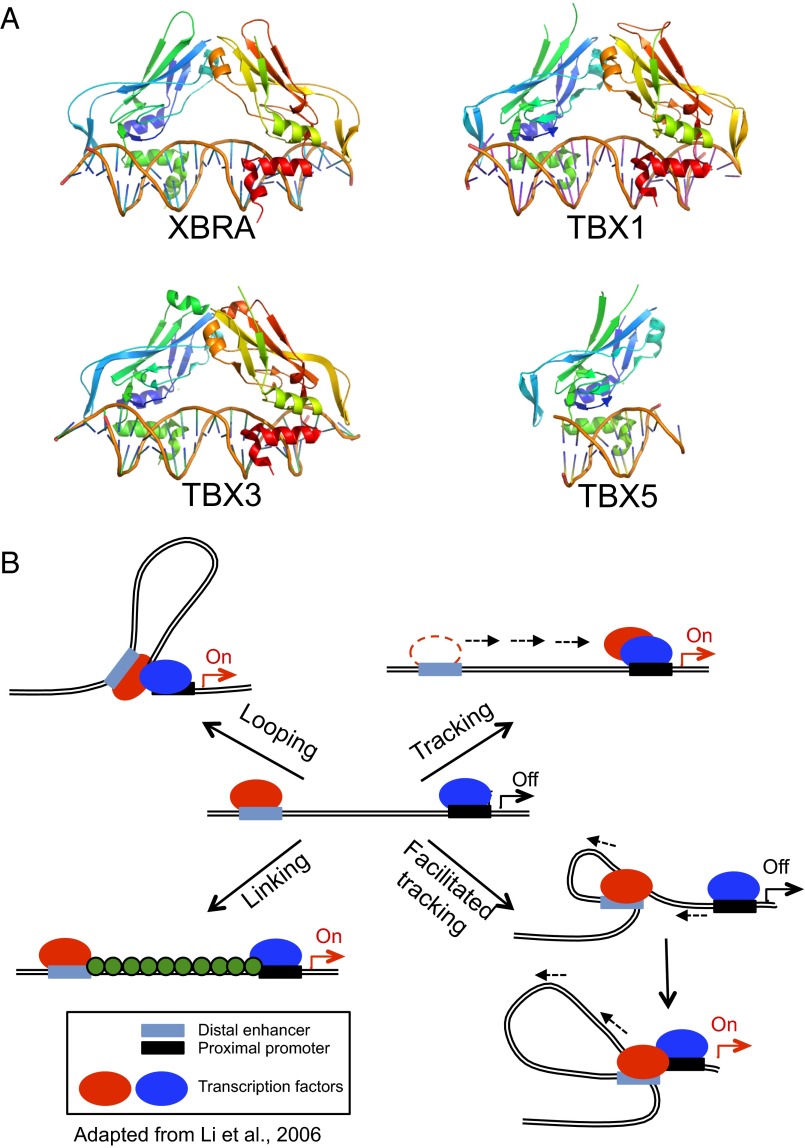
The Tbox transcription factors bind contiguous DNA elements. (A) Structures of the Tbox DBD–DNA complexes of Xbra (PDB ID code 1XBR), TBX1 (PDB ID code 4A04), TBX3 (PDB ID code 1H6F), and TBX5 (PDB ID code 2X6V). Note that the overall folds of the four Tbox domains are essentially identical, as is their mode of recognition of the B-form DNA double helix. Xbra, TBX1, and TBX3 crystallized as homodimers and bind two consensus target sequences, respectively, but the dimers are not tightly associated and probably bind their two sites independently; TBX5 binds as a monomer to a single site. (B) Models for action at a distance by a dimeric transcription factor. The two subunits of the dimer are shown in red and blue and may be either different (heterodimer) or identical (homodimer). In a looping model, each subunit binds independently to a promoter recognition element and a distant element, which may be either another promoter or, as shown, an enhancer sequence. Formation of the dimer juxtaposes the distant sites. Tracking postulates that the different elements serve to recruit the protein subunits to the DNA, but that one of them then scans along the chromosome, leaving its site behind, until it encounters its cognate partner. Facilitated tracking combines elements of tracking and looping, in that the distant site is brought along with its binding protein. In a linking model, the two subunits and their sites remain separate and other proteins (shown in green) connect them. The previously known Tbox protein structures shown in A are either monomeric or bind contiguous DNA elements and consequently would not be expected to act at a distance as depicted in these models.