Fig. 4. Sensitivity analysis.
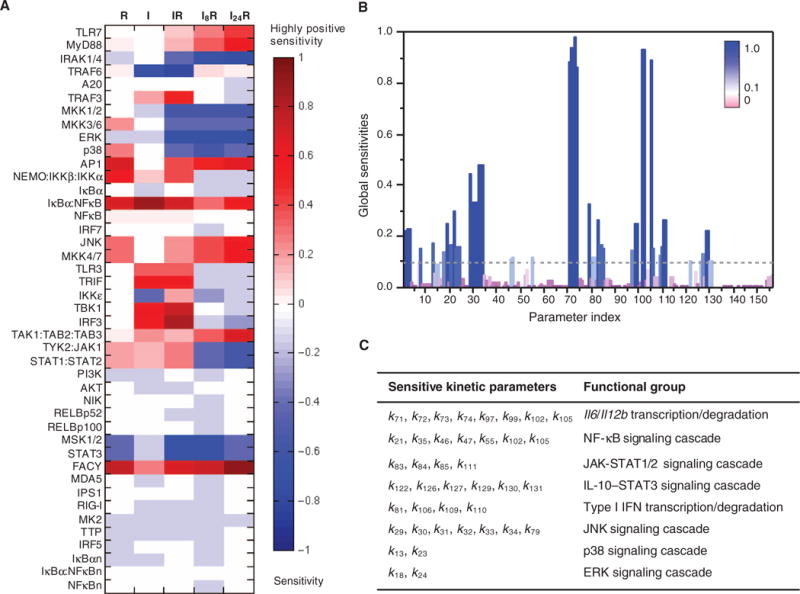
(A) Local sensitivities (control coefficients) of the initial concentrations of proteins (listed along the ordinate) to the integrated response of Il12b expression under the R, I, IR, I8R, and I24R conditions. The heat map is color-coded from red (high positive sensitivity) to blue (high negative sensitivity). The former group refers to proteins whose increase in initial concentration would enhance cytokine mRNA production, and the latter group refers to those whose decrease in initial concentration might enhance cytokine mRNA production. Components whose initial concentrations had little or no effect on the system dynamics are shown in light colors or white. (B) Global sensitivities calculated according to the SMC-MPSA method. Sensitive parameters, which strongly influenced the observed behavior, are depicted as dark blue peaks. Robust parameters, whose variation had little effect on the model dynamics, are depicted in purple. The highest peaks refer to transcription or degradation rate constants for Il6 and Il12b mRNAs. (C) Functional classification of the 40 kinetic parameters that led to the highest sensitivity peaks, which are those above the dotted threshold line in (B). The corresponding reactions are indicated by red arrows in fig. S1.
