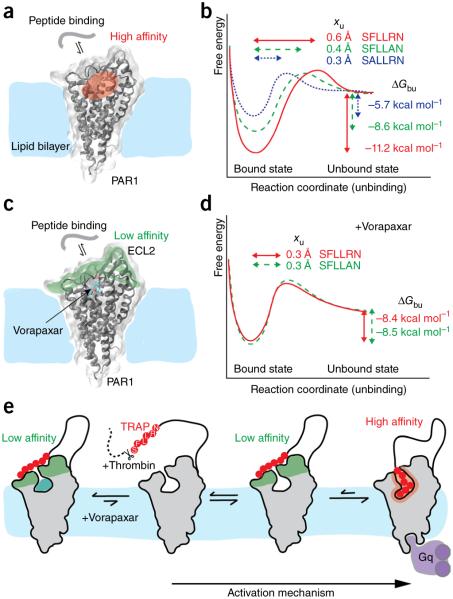
Figure 5.
Free-energy landscape describing the thermodynamic (ΔGbu) and kinetic (xu) parameters of peptide-based ligands binding to PAR1. (a) Cartoon showing a peptide-based ligand binding to PAR1 through a high-affinity binding site, which is expected to be in the region shaded red3. (b) Free-energy binding landscape of three different peptide-based ligands depends on the sequence of the peptide. xu represents the distance to the transition state separating the ligand-bound and unbound state and is indicated for each peptide by horizontal arrows. ΔGbu gives the free-energy difference between the ligand-bound and unbound states and is indicated for each peptide by vertical arrows. (c) Cartoon showing a peptide-based ligand interacting to vorapaxar-bound PAR1 through a low-affinity binding site, which is expected to be in the region shaded green. (d) Free-energy landscape of ligands binding to vorapaxar-bound PAR1. PAR1 structures are shown in the vorapaxar-bound state (PDB ID 3VW7). (e) Binding model of the native SFLLRN ligand (red) to PAR1. For both the vorapaxar-inhibited and unbound states of PAR1, the ligand binds at low affinity to the extracellular PAR1 surface, from which extracellular loops 2 and 3 have been proposed to bind the ligand3. In the presence of the antagonist vorapaxar, the native ligand cannot bind to the high-affinity binding site (or state). In the absence of vorapaxar, the native ligand can bind the high-affinity site; this functionally activates PAR1, leading to the binding of Gq, a class of G proteins that participate in a variety of cellular signaling pathways.