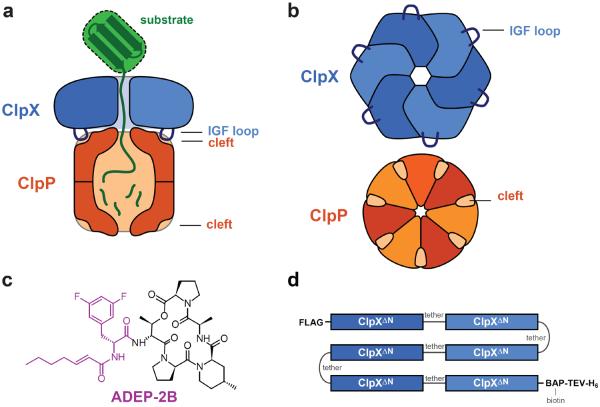
Figure 1.
The ClpXP protease. a) Side view of ClpXP degrading a substrate (green). A ClpX hexamer (blue) recognizes, unfolds, and translocates protein substrates into the degradation chamber of ClpP (dark orange), which consists of two heptameric rings. ClpXP is principally stabilized by interactions between the IGF loops of ClpX and hydrophobic clefts on each ClpP ring. b) Axial view of a ClpX homohexameric ring and a ClpP homoheptameric ring, highlighting the interaction elements. c) Chemical structure of ADEP-2B.21,28 The portion thought to mimic binding of an IGF tripeptide is colored purple. d) sc6ClpXΔN-bio is a single-chain pseudohexamer in which the ClpXΔN subunits are linked by six-residue tethers. The protein contains an N-terminal FLAG tag and a C-terminal sequence consisting of a biotin acceptor peptide (BAP), a cleavage site for Tobacco Etch Virus protease (TEV), and six histidines (H6).