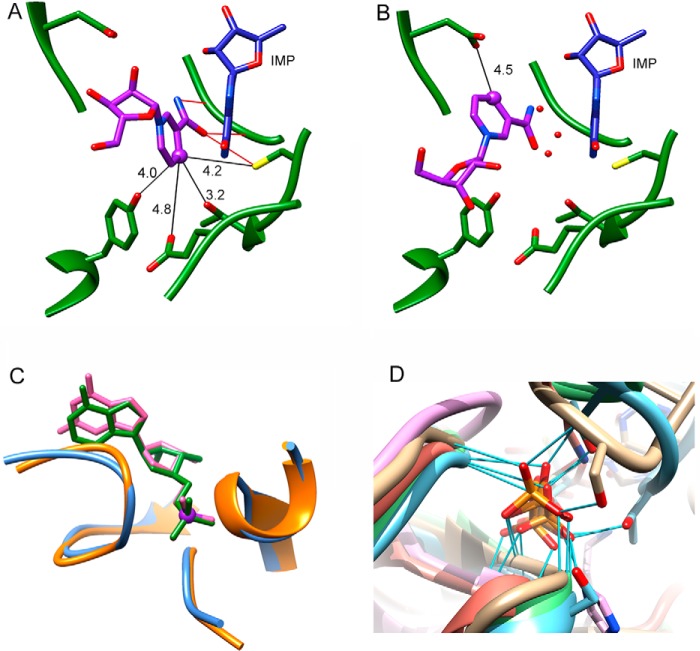
FIGURE 2.
Interactions of substrate and cofactors with GMPR. A, interaction of nicotinamide riboside (purple) in the IN conformation (PDB code 2C6Q; subunit B). Residues within 5 Å of nicotinamide are shown (the diphosphates and 2′-phosphoadenosine are omitted for clarity). The distances are depicted by black lines and have Å units. Nicotinamide hydrogen bonds are presented by red lines. IMP is depicted in blue, and GMPR is shown in green. B, interactions of the nicotinamide riboside portion of NADP+ (purple) in the OUT conformation (PDB code 2C6Q; subunit A). The red spheres represent additional water molecules that were found in the OUT conformation within 3.5 Å of inosine. C, substrate phosphate group binding in GMPR. Structure and interaction of phosphate group in E·GMP (backbone in orange and GMP in magenta; PDB code 2A7R) and E·IMP·NADPH (backbone in steel blue and IMP in green; PDB code 2C6Q) crystal structures. D, superposition of the standard phosphate binding site in (β/α)8 barrel proteins. Representatives of five different (β/α)8 superfamilies are shown (2): GMPR (2C6Q, chain A) (tan); thiamine phosphate synthase (2TPS, chain A) (blue); quinolinate phosphoribosyltransferase (1QPR, chain A) (pink); triose-phosphate isomerase (1TPH, chain 1) (green); and ribulose-bisphosphate carboxylase/oxygenase (1RBL, chain A) (orange). The hydrogen bonds between the phosphate group and the protein residues are depicted by light blue lines. This figure was rendered with UCSF Chimera (27).