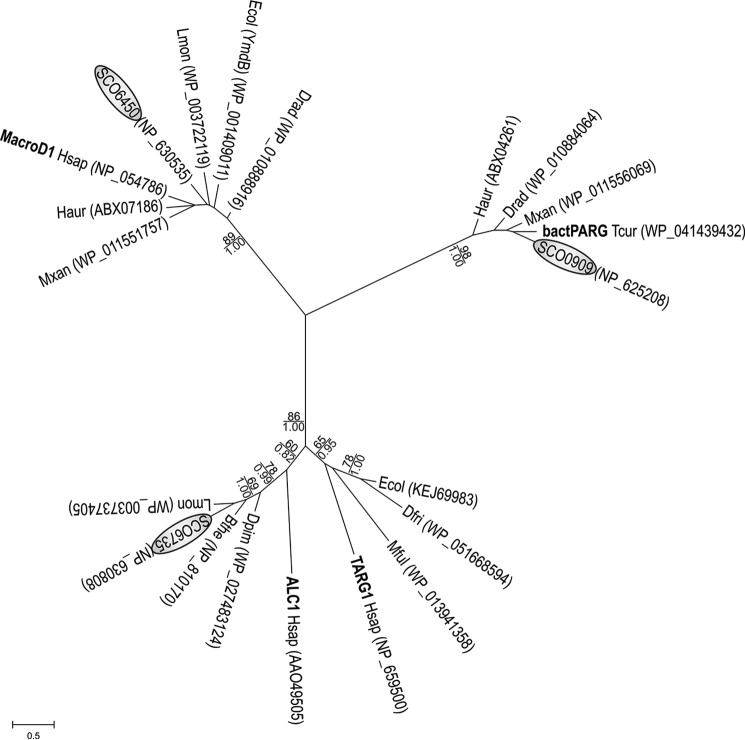
FIGURE 1.
Maximum likelihood phylogenetic tree illustrating the relationship between selected representative macrodomains. Human proteins (Hsap) and bacterial PARG from Thermomonospora curvata (Tcur) are shown in bold type. S. coelicolor proteins are circled. Remaining macrodomain proteins were selected from the following bacterial species: B. thetaiotaomicron (Bthe), Deinococcus frigens (Dfri), Deinococcus pimensis (Dpim), D. radiodurans (Drad), E. coli (Ecol), Herpetosiphon aurantiacus (Haur), Listeria monocytogenes (Lmon), Myxococcus fulvus (Mful), and M. xanthus (Mxan). Sequence accession numbers are given in parentheses. Bootstrap values ML (>60%) are given above the lines, and MCMC values are given (>0.6) below the lines. The scale bar indicates the genetic distance of the branch lengths.