Fig. 4.
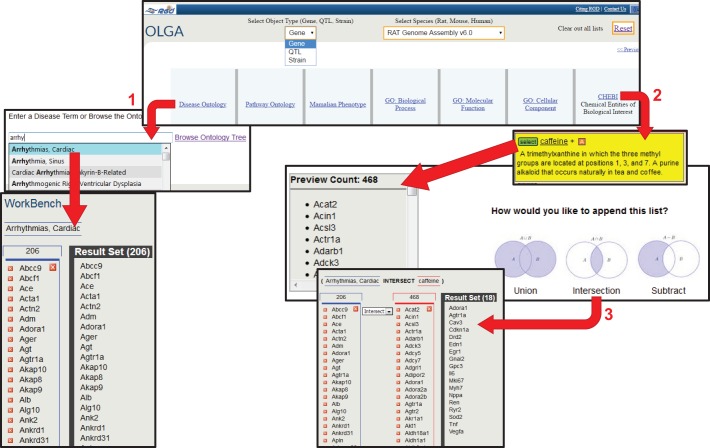
The Object List Generator and Analyzer (OLGA) tool facilitates construction of complex queries for rat, mouse or human genes or QTLs, or rat strains. In the example displayed, two queries are made for rat genes based on their functional annotations. In the first step (1), the Disease Ontology is searched. As the user types, an autocomplete list of disease terms is shown and the term “Arrhythmias, Cardiac” was selected. The resulting list, shown in the ‘WorkBench’ section of the page with the term used for the search, contains 206 genes. In the second step (2), the CHEBI ontology was browsed to locate the term “caffeine”. Selecting this term returns a preview list of 468 genes. Three options are given for appending the second list onto the first: ‘Union’, which combines the lists to produce the total non-redundant set of genes found in either list; ‘Intersection’, which returns the list of genes in common between the two; and ‘Subtract’, which returns only the genes from the first list that do not appear in the second. ‘Intersection’ was selected in this example (3) to produce a set of 18 genes that are associated with cardiac arrhythmias and interact with caffeine, with annotations. Accessed 15 April, 2016.