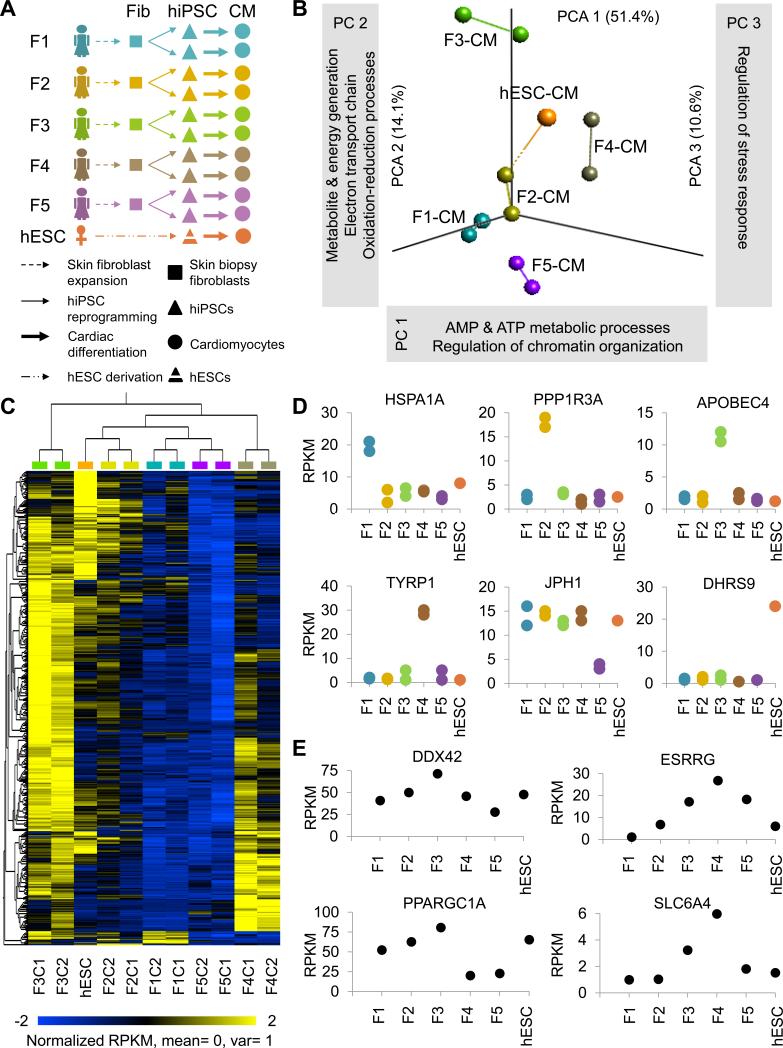
Figure 1. Inter- and intra-patient variation analysis in hiPSC-CMs. See also Figures S1 and S2, and Tables S1-S4.
(a) Schematic outline of study design. Skin fibroblasts were collected from 5 female volunteers, reprogrammed to hiPSCs, and two hiPSC clones per patient were differentiated to CMs for RNA sequencing, pathway analysis, and drug testing. (b) Principal component analysis (PCA) with computation of closest neighboring samples and (c) hierarchical clustering showing closer correlation between hiPSC-CMs from hiPSC clones of the same individual (intra-patient correlation), rather than between individuals (inter-patient variation). (d) The most highly differentially expressed genes for each line were conserved in hiPSC-CMs from both hiPSC clones analyzed. (e) Gene ontology (GO) clusters least conserved in individuals’ hiPSC-CMs were associated with ATP metabolism and chromatin organization (PC1), redox reactions and the electron transport chain (PC2), and regulation of stress response (PC3). Among the most highly differentially expressed genes for each principal component were DDX2 (PC1), ESRRG (PC1), PPARGC1A (PC2), and SLC6A4 (PC3).