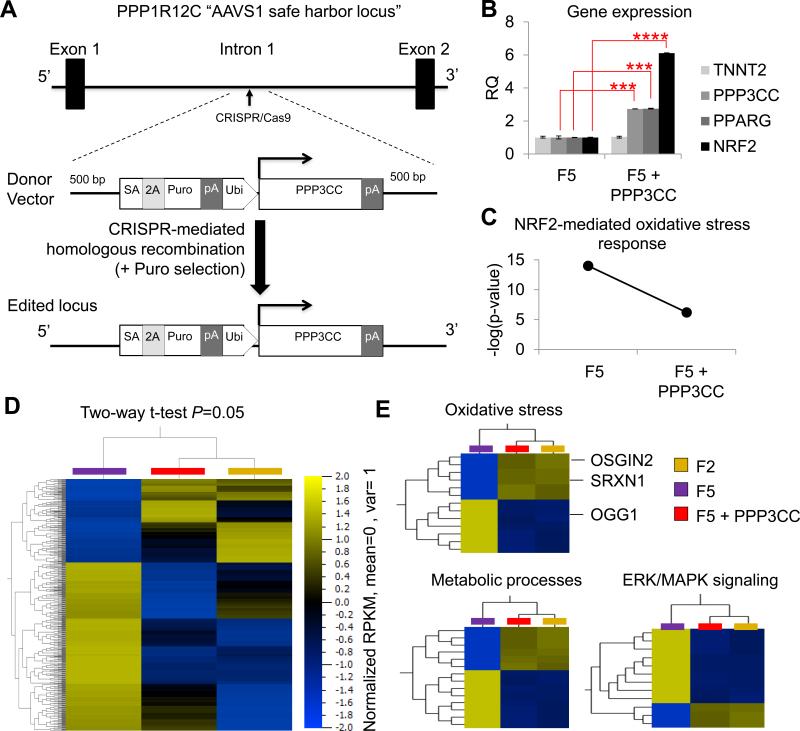
Figure 6. Generation and characterization of genome-edited F5 hiPSC-CMs. See also Figure S7 and Tables S5-S7.
(a) Schematic diagram of the CRISPR/Cas9 experimental strategy used to over-express the PPP3CC gene in the AAVS1 safe harbor locus of F5 hiPSCs. (b) Quantitative real-time PCR analysis of PPP3CC, PPARG, NRF2, and TNNT2 gene expression in F5 hiPSC-CMs and CRISPR/Cas9-mediated genome edited F5 hiPSC-CMs overexpressing PPP3CC (F5 + PPP3CC) from the ubiquitin promoter inserted in the AAVS1 safe harbor locus. (c) Pathway analysis for prediction of risk towards cytotoxicity revealed a reduced probability for NRF2-mediated oxidative stress in F5 + PPP3CC hiPSC-CMs compared to parental F5 hiPSC-CMs. (d) Hierarchical clustering of AmpliSeq RNA-sequencing data under two-way t-test (P = 0.05) showed that many genes had similar gene expression levels in F2 and genome edited F5 + PPP3CC hiPSC-CMs, while F5 hiPSC-CMs had a distinctly different gene expression pattern. (e) Gene ontology (GO) analysis identified that the most differentially expressed genes were involved in metabolic processes, oxidative stress, and signal transduction (e.g., ERK/MAPK signaling). SA, splice acceptor; Puro, puromycin resistance gene; Ubi, ubiquitin promoter; pA, polyadenylation signal.