Figure 1.
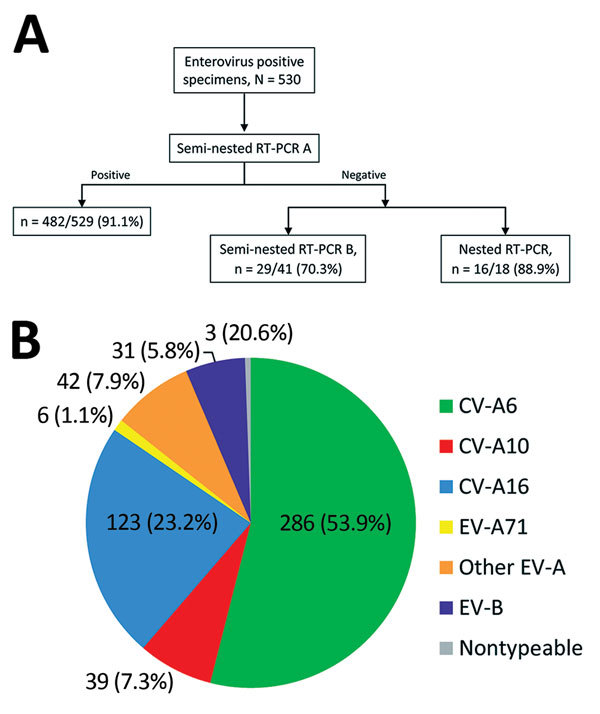
Methodologic approach for enterovirus genotyping and distribution of types associated with hand, foot and mouth disease and herpangina, France, April 2014–March 2015. A) Semi-nested reverse transcription PCR (RT-PCR) A using primers specifically developed for enterovirus types belonging to the EV-A species was first performed for all clinical samples except 1. For this sample, the viral load was low, and the nested RT-PCR described by Nix et al. (27) was performed directly. If the semi-nested RT-PCR A was negative, the genotyping was alternatively performed by a semi-nested RT-PCR B with primers specific to the EV-B species (28) or a nested RT-PCR (27). B) Among other EV-A species, 5 different types were identified: coxsackievirus (CV) A4, n = 18; CV-A8, n = 16; CV-A2 and CV-A5, n = 5 each; and CV-A12, n = 1. Among EV-B species, 12 different types were identified: echovirus (E) 16 (E-16) and E-18 (n = 5 each); E-11 and coxsackievirus B3 (CV-B3; n = 4 each); CV-B1, CV-B2, CV-B4, CV-A9, and E-6 (n = 2 each); and E-3, E-5, and E-25 (n = 1 each). EV-A, Enterovirus A; EV-A71, enterovirus A71; EV-B, Enterovirus B; RT-PCR, reverse transcription PCR.
