Fig. 1.
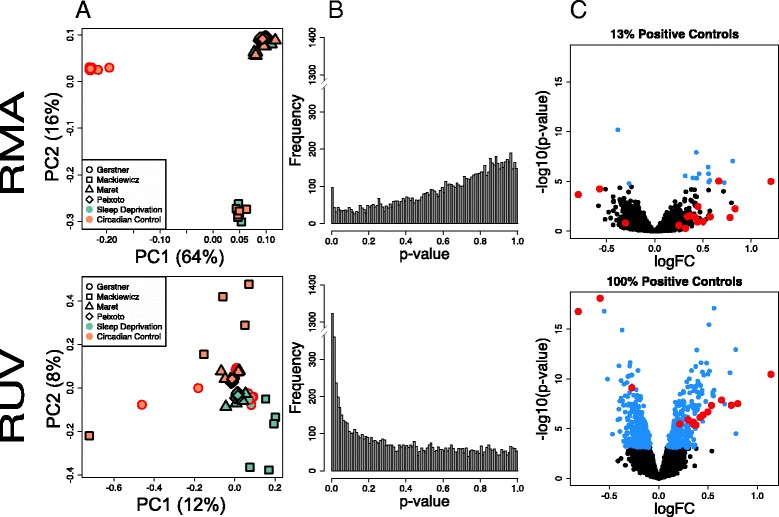
Integrated analysis of the effect of 6 h of sleep deprivation in the murine cortex. a Scatterplots of first two principal components (PC, log-scaled, centered intensities) following RMA and RUV normalization. Percent variance explained by each PC in parenthesis. Triangles denote samples from the Maret et al. 2007 study [2] (whole brain), square samples from the Mackiewicz et al. 2007 study [3] (cortex), rhombus the samples from Vecsey, Peixoto et al. 2012 [8] (hippocampus), and circles are samples from this study (cortex). In green, samples following 6 h of sleep deprivation (SD); in orange, time of day matched controls (CC). Samples cluster according to array platform (PC1) and lab (PC2) following Quantile normalization. After applying RUV, samples cluster according to treatment (PC2). b Distribution of unadjusted p-values for tests of differential expression between SD and CC samples following Quantile and RUV normalization. The distribution of p-values following Quantile normalization is not uniform and biased towards 1. RUV returns uniformity to the p-value distribution and increases discovery of differentially expressed genes (genes that have a low p-value). c Volcano plot of differential expression (−log10 p-value vs log fold change) of Quantile and RUV normalized samples. Genes with an FDR <0.01 are highlighted in blue. Positive controls are circled in red; RUV increases the detection of known differentially expressed genes from 0 to 100 %. PCA plots were performed using the R/Bioconductor package EDASeq (v. 2.0.0). RUVs normalization was performed using the R/Bioconductor package RUVSeq (v. 1.0.0). Differential expression analysis was performed using R/Bioconductor package limma
