Fig. 1.
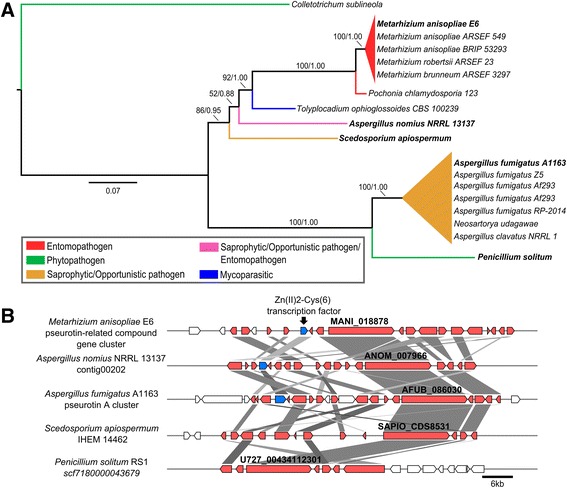
Pseurotin-related compound BGC (MaNRPS-PKS2). a Phylogenetic analysis was performed using Maximum-likelihood and Bayesian methods, based on the pseurotin-related backbone gene and orthologous sequences exhibited by several fungi. The orthologous sequences were classified according to fungal lifestyle trait, represented by different colors. The Bayesian tree is displayed, and branch support values (bootstrap proportions and Bayesian posterior probability) are associated with nodes. The Bayesian inference ran for 9,997,000 generations. Species in bold in (a) were used for the cluster conservation analysis presented in (b). b Some genes from M. anisopliae MaNRPS-PKS2 BGC resembled the characterized pseurotin BGC from A. fumigatus (34–81 % identity) and putative BGCs from A. nomius (49–85 % identity), S. apiospermum (63–84 % identity) and P. solitum (59–81 % identity). The M. anisopliae Zn(II) 2-Cys(6) transcription factor resembles the embedded transcription factor found in A. fumigatus (34 % identity), and the putative transcription factor from A. nomius (49 % identity). Interestingly, S. apiospermum and P. solitum do not have orthologs for this transcription factor. Orthologous genes were assigned the same color; white boxes represent genes that were not predicted to be part of M. anisopliae cluster, and blue boxes represent the conserved Zn(II)2-Cys(6) transcription factor
