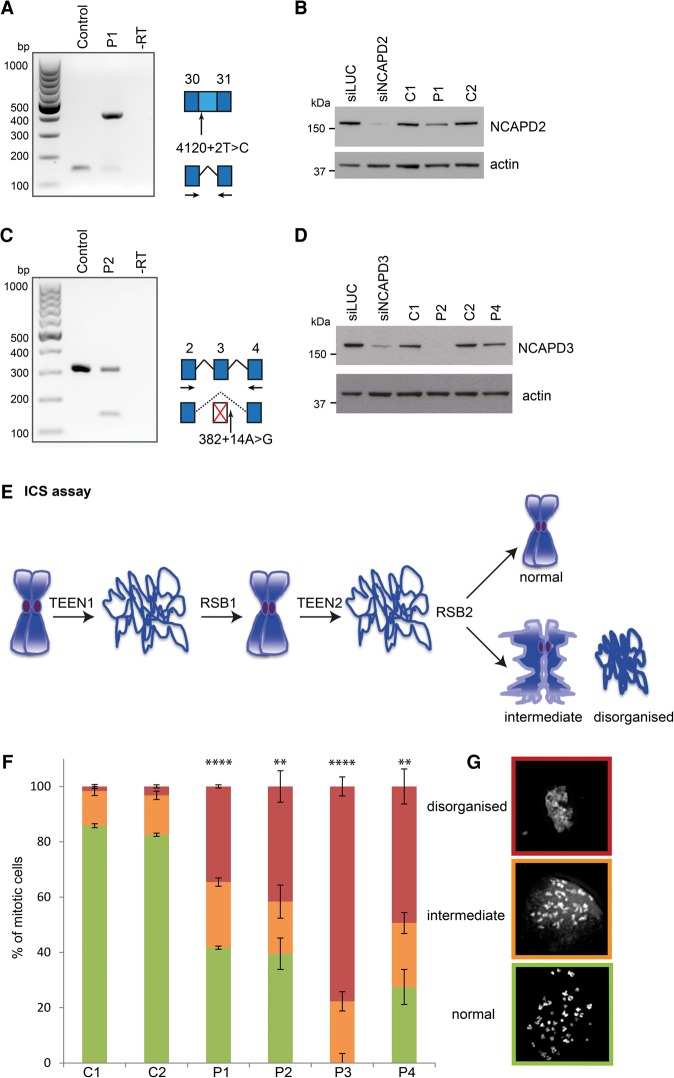
Figure 2.
Mutations impair canonical condensin function in mitotic chromosome compaction. (A,B) The c.4120+2T>C mutation results in retention of intron 30 in NCAPD2 transcripts and reduced NCAPD2 protein levels. (A) RT–PCR using primers in exons 30 and 31 (arrows in the schematic) demonstrated altered splicing of NCAPD2 in patient P1 fibroblasts. The larger 413-base-pair (bp) PCR product corresponded to a transcript containing intron 30, confirmed by subcloning and Sanger sequencing. This transcript encodes an alternative C terminus comprising an additional 29 amino acids before a PTC with consequent loss of 28 amino acids encoded by exon 31. (B) NCAPD2 protein levels are reduced in P1 fibroblasts. Immunoblot with anti-NCAPD2 antibody. (Left two lanes) RNAi of NCAPD2 in RPE1 cells confirms the specificity of the NCAPD2 antibody. (Remaining lanes) Patient P1 and control primary fibroblasts. NCAPD2 protein mobility is not expected to be altered in patient P1, as the mutant protein has a molecular weight similar to that of wild type. (Bottom panel) Loading control: The blot was probed with anti-actin antibody. (C,D) The NCAPD3 c.382+14A>G mutation leads to skipping of exon 3, which, in combination with c.1783_1784delG frameshift mutation in trans, results in markedly reduced NCAPD3 protein. (C) Alternative splicing of NCAPD3 in patient P2 fibroblasts, detected by RT–PCR using primers in exon 2 and exon 4 (arrows, schematic). The wild-type transcript represented by a PCR product of 317 bp is substantially reduced in patient P2 cells, while the smaller PCR fragment of 154 bp is detectable only in patient P2 and corresponds to a transcript in which exon 3 had been skipped (confirmed by subcloning and sequencing). This results in an out-of-frame transcript, which is expected to be subject to NMD. (D) Immunoblotting of NCAPD3 establishes that protein levels are markedly reduced in patient P2. (Left two lanes) RNAi of NCAPD3 in RPE1 cells demonstrating specificity of the NCAPD3 antibody. (Remaining lanes) Patient and control primary fibroblasts. NCAPD3 protein is substantially depleted in patient P2, whereas protein levels are unaffected by the p.Glu1153Ala missense mutation in patient P4 fibroblasts. (Bottom panel) Loading control: The blot was probed with anti-actin antibody. (E) Schematic of intrinsic chromosome structure (ICS) assay. Sequential TEEN decompaction and refolding in RSB buffer was used to establish the structural integrity of mitotic chromosomes. (F,G) Patient mutations lead to compromised ICS. (F) Quantification of abnormal chromosome refolding after ICS assay in C1 and C2 and patient P1–P4 cell fibroblasts (experiments = 3; n > 30 mitoses per sample per experiment). Error bars indicate SEM. Two-tailed t-test, (**) P ≤ 0.01; (****) P ≤ 0.0001; proportion of “disorganized” chromosomes versus C2. (G) Representative images of chromosome morphology from ICS assay. Chromosomes were visualized with DAPI following two sequential rounds of decompaction (TEEN) and refolding (RSB). Patient and control primary fibroblast chromosomes were scored as either reforming the original chromosome morphology (“normal”), partially reforming starting chromosome morphology (“intermediate”), or complete loss of chromosome compaction (“disorganized”).