In this study, Subasic et al. investigated the post-transcriptional control of caspases. The authors describe four conserved RNA-binding proteins (RBPs) that sequentially repress the CED-3 caspase in distinct regions of the C. elegans germline and identify seven RBPs that regulate human caspase-3 expression and/or activation, suggesting that translational inhibition of executioner caspases by RBPs might be a general strategy used widely across the animal kingdom to control apoptosis.
Keywords: CED-3, GLD-1, RNA-binding proteins, apoptosis, caspase, caspase-3, translational control
Abstract
Caspases are key components of apoptotic pathways. Regulation of caspases occurs at several levels, including transcription, proteolytic processing, inhibition of enzymatic function, and protein degradation. In contrast, little is known about the extent of post-transcriptional control of caspases. Here, we describe four conserved RNA-binding proteins (RBPs)—PUF-8, MEX-3, GLD-1, and CGH-1—that sequentially repress the CED-3 caspase in distinct regions of the Caenorhabditis elegans germline. We demonstrate that GLD-1 represses ced-3 mRNA translation via two binding sites in its 3′ untranslated region (UTR), thereby ensuring a dual control of unwanted cell death: at the level of p53/CEP-1 and at the executioner caspase level. Moreover, we identified seven RBPs that regulate human caspase-3 expression and/or activation, including human PUF-8, GLD-1, and CGH-1 homologs PUM1, QKI, and DDX6. Given the presence of unusually long executioner caspase 3′ UTRs in many metazoans, translational control of executioner caspases by RBPs might be a strategy used widely across the animal kingdom to control apoptosis.
The elimination of unwanted cells by apoptosis is indispensable for animal development and tissue homeostasis. Dysregulation of apoptosis has been linked to a number of diseases, including autoimmune and cardiovascular disorders, neurodegeneration, and cancer (Fuchs and Steller 2011). Much of the mechanism by which cells undergo apoptosis was unraveled in early studies in Caenorhabditis elegans, in which it became clear that the process is evolutionarily conserved. In all species analyzed, activation of caspases is the last step of the canonical apoptotic cascade (Supplemental Fig. S1; Conradt 2009). Due to the detrimental consequences of inappropriate cell death activation, caspases are regulated at multiple levels. In addition to transcriptional control, caspases are subject to significant post-translational regulation, in particular through proteolytic processing (Earnshaw et al. 1999). Caspases are also regulated by caspase-like decoy molecules that can bind and sequester procaspase zymogens or compete for activation with caspases (Lamkanfi et al. 2007). In Drosophila and mammals, inhibitors of apoptosis (IAPs) can directly inhibit activated caspases (Gyrd-Hansen and Meier 2010), and, in C. elegans, CSP-2 and CSP-3 have been found to inhibit autoactivation of CED-3 in the germ cells and soma, respectively (Geng et al. 2008, 2009). However, relatively little is known about the potential regulation of caspases at the post-transcriptional level.
Results
Four conserved RNA-binding proteins (RBPs) sequentially repress the CED-3 caspase in the C. elegans germline
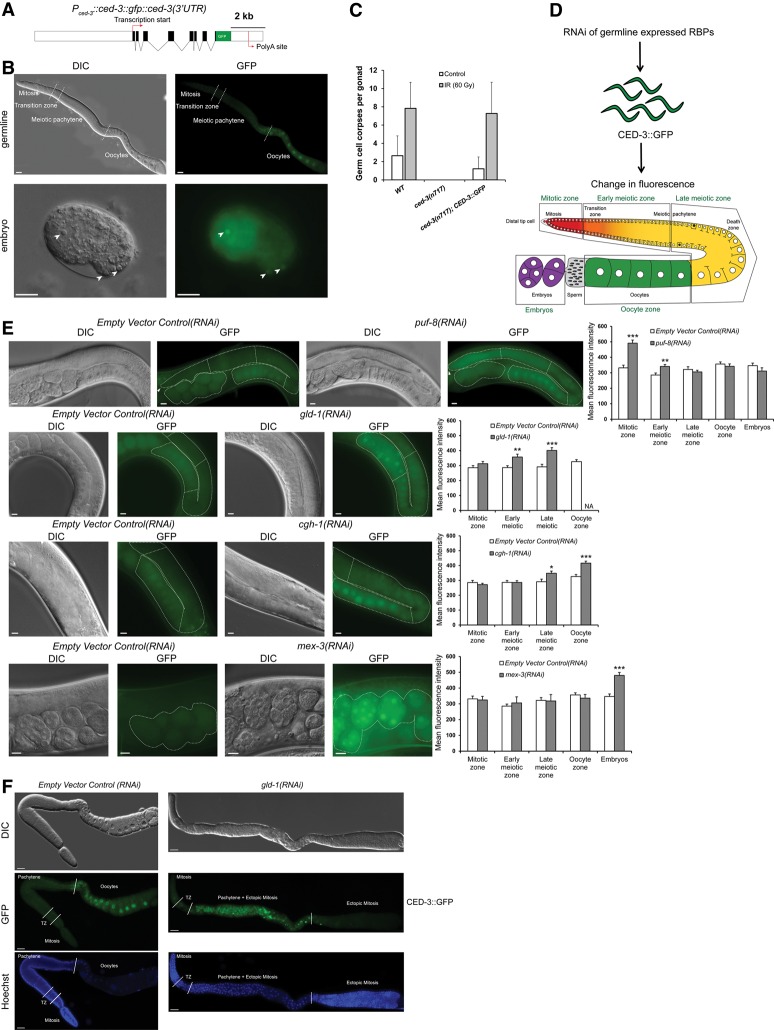
To examine CED-3 expression in C. elegans and generate a screening tool for identifying molecules that may regulate its expression, we generated a transgenic strain expressing a C-terminally GFP-tagged CED-3 under its endogenous promoter and 3′ control region {opIs461[Pced-3::ced-3::gfp::ced-3(3′ UTR)]} (Fig. 1A,B). This CED-3::GFP translational reporter was expressed mostly in embryos and the adult germline, where extensive apoptosis is known to occur (Gartner et al. 2008; Conradt 2009). The GFP-tagged protein could fully rescue the absence of physiological and DNA damage-induced germ cell apoptosis in ced-3(n717) mutants (Fig. 1C). In embryos, we often found an enrichment of GFP signal within the apoptotic corpses compared with the surrounding cells (Fig. 1B), consistent with two previous studies (Maurer et al. 2007; Chakraborty et al. 2015). In the germline, CED-3::GFP was expressed primarily in the oocyte region, with very weak to no expression in the mitotic and early meiotic zones. In oocytes, CED-3::GFP showed a significant enrichment in the nuclei, whereas, in the rest of the germline, we observed an accumulation of GFP signal in the perinuclear region (Supplemental Fig. S2).
Figure 1.
RNAi screen for CED-3 regulators identified four conserved RBPs (PUF-8, GLD-1, CGH-1, and MEX-3) that negatively regulate CED-3 expression in different regions of the germline. (A) Structure of the reporter fusion with GFP [Pced-3::ced-3(genomic)::gfp::ced-3(3′ UTR)] used in this study. The promoter and 3′ untranslated region (UTR) elements are marked as white boxes, exons are marked as black boxes, and introns are marked as lines. (B) The CED-3 translational reporter (CED-3::GFP) shows germline and embryonic expression. Differential interference contrast (DIC) and GFP images of an adult germline and an embryo expressing CED-3::GFP are shown. White arrowheads indicate apoptotic corpses in the embryo enriched in CED-3 expression. Bar, 10 µm. (C) The CED-3::GFP translational reporter rescues the absence of physiological and irradiation-induced germ cell death in ced-3(n717) mutants. Error bars represent SD from two independent experiments. n = 25 animals per experiment. (D) Scheme of RNAi screen via feeding used to identify germline-expressed RBPs that regulate CED-3 expression. Four different germline areas (the mitotic, early meiotic, late meiotic, and oocyte zones) and fertilized embryos were monitored for change in fluorescence. (E) DIC and GFP images of the four positives from the screen that altered CED-3 expression: PUF-8 in the mitotic and early meiotic zones, GLD-1 in the early and late meiotic zones, CGH-1 in the late meiotic and oocyte zones, and MEX-3 in embryos. Empty vector control(RNAi) was used as a control. Average mean GFP fluorescence intensity was quantified in the five zones in 10–20 animals. Dashed lines outline the gonads, and arrowheads indicate the positions of the distal tip cells. Different zones selected for quantification (as in D) are separated with straight dashed white lines. Error bars represent SEM. n ≥ 10. Asterisks represent the P-value for a paired t-test comparing the reporter GFP intensity in RBP(RNAi)-treated and empty vector control(RNAi)-treated animals. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Bar, 10 µm. (F) DIC, GFP, and Hoechst images of dissected gonads show increased CED-3::GFP expression in the meiotic pachytene region but not in the tumorous ectopic proliferative zone of fully depleted gld-1(RNAi) animals carrying the CED-3::GFP reporter. Bar, 10 µm. See also Supplemental Figures S2 and S4.
The C. elegans germline is an exceptionally useful tissue to study apoptosis. Germ cells can activate apoptosis in response to developmental signals as well as a wide range of external cues, including DNA damage, bacterial infection, and the failure of meiotic chromosomes to synapse (Bailly and Gartner 2013). The C. elegans germline relies extensively on translational control to regulate gene expression (Merritt et al. 2008). To test whether germline CED-3 expression might be controlled at the translational level, we performed a targeted RNAi screen with 20 RBPs known to be expressed in the germline (Fig. 1D; Supplemental Table S1). Several of the candidates, including gla-1, cpb-3, car-1, cgh-1, and gld-1, were reported previously to inhibit apoptosis in the C. elegans germline (Lettre et al. 2004; Boag et al. 2005; Schumacher et al. 2005; Kritikou et al. 2006). RNAi of four of the tested RBPs strongly increased CED-3::GFP expression in distinct regions of the germline: puf-8 in the mitotic and early meiotic zones, gld-1 in the early and late meiotic zones, cgh-1 in the late meiotic zone, and oocytes and mex-3 in embryos (Fig. 1E). The regions with altered CED-3::GFP expression were consistent with the previously determined expression pattern of the four RBPs: PUF-8 is highly expressed in mitotic cells but low in meiotic pachytene and oocytes (Racher and Hansen 2012), GLD-1 is low to absent in mitotic cells and oocytes but high in meiotic pachytene (Jones et al. 1996; Schumacher et al. 2005), and MEX-3 is high in oocytes and enriched in the anterior fated cells in the embryo (Draper et al. 1996), while CGH-1 is essentially ubiquitous throughout the germline and early embryo before becoming enriched in the P lineage (Navarro et al. 2001).
MEX-3 is a KH domain RBP required for embryonic cell fate specification and maintenance of germline totipotency (Draper et al. 1996; Ciosk 2006). mex-3 mutants produce excess muscle, fail to develop proper body morphology, and consequently die as embryos. PUF-8 belongs to the group of Pumilio family of RBPs that promotes germline stem cell mitosis redundantly with MEX-3 (Ariz et al. 2009). Animals lacking PUF-8 form germ cell tumors due to hyperproliferation of dedifferentiated primary spermatocytes (Subramaniam and Seydoux 2003). CGH-1 is a conserved DEAD-box germline helicase required for proper oocyte development and is linked to germ cell apoptosis (Boag et al. 2005; Tomazella et al. 2012). GLD-1 is a KH domain RBP that belongs to the conserved STAR family and is a master regulator of translation of several hundred transcripts in the C. elegans germline (Lee and Schedl 2001; Jungkamp et al. 2011; Wright et al. 2011). GLD-1 is known to inhibit germline apoptosis via translational repression of the C. elegans p53 tumor suppressor homolog cep-1 (Schumacher et al. 2005).
GLD-1 represses ced-3 mRNA translation via two binding sites in its 3′ untranslated region (UTR)
In strong gld-1 knockdown and null mutants, germ cells fail to complete meiosis and re-enter the mitotic cell cycle, resulting in an ectopic proximal proliferation zone (Francis et al. 1995; Lee and Schedl 2001). We stained the DNA of dissected gonads from fully depleted gld-1(RNAi) animals with Hoechst to differentiate between mitotic and meiotic cells in the germline and found increased CED-3::GFP expression in the central meiotic region of the germline where GLD-1 is normally expressed but not in the distal mitotic or ectopic proliferative zone (Fig. 1F). Temperature-sensitive gld-1(op236) mutants raised at 25°C recapitulated the RNAi results, showing a strong increase of CED-3::GFP expression in the pachytene region of the germline (Supplemental Fig. S4C).
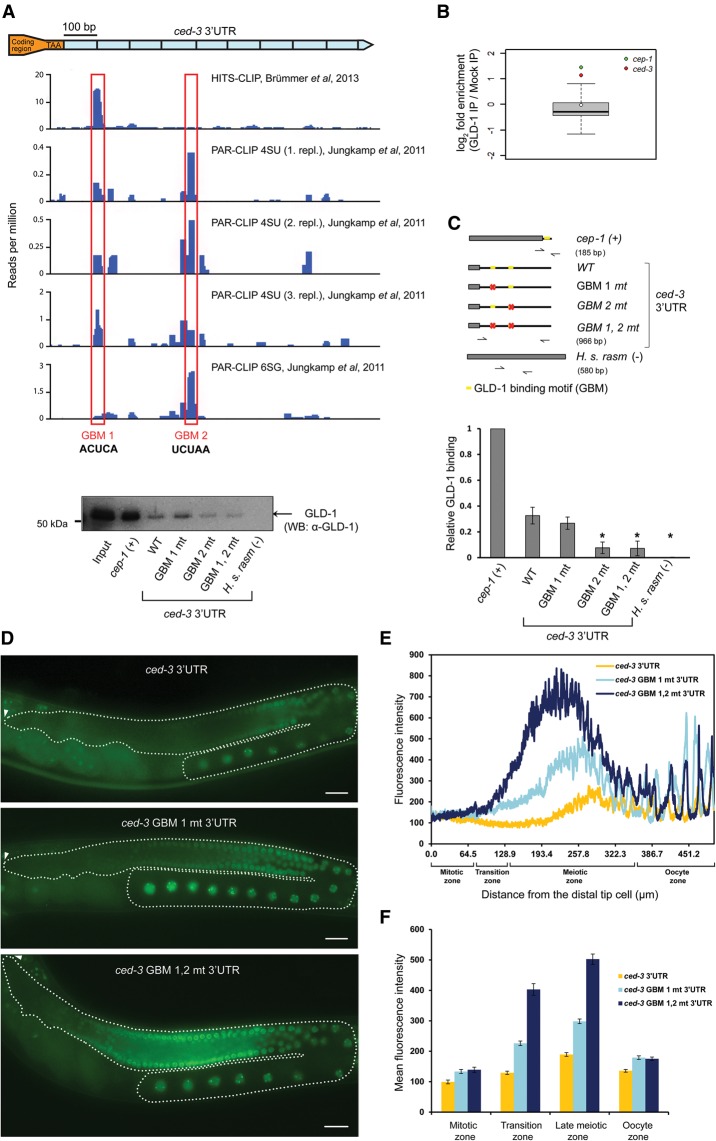
In order to test whether GLD-1 binds directly to the ced-3 mRNA, we reanalyzed a set of transcriptome-wide GLD-1-binding sites that we generated previously by high-throughput sequencing of RNA isolated by cross-linking and immunoprecipitation (HITS-CLIP) of a transgenic line expressing a rescuing GLD-1::STREP/HA transgene (Brümmer et al. 2013). We identified an enrichment of clipped reads at one site in the ced-3 3′ UTR (Fig. 2A). This site (GMB 1) contained a sequence that matches the GLD-1-binding motif (GBM) that we inferred previously using motif-based enrichment of the reads from the HITS-CLIP experiment (Brümmer et al. 2013). Inspection of an additional published data set of GLD-1-binding sites generated via photoactivatable ribonucleoside-enhanced CLIP (PAR-CLIP) (Jungkamp et al. 2011) identified evidence for a second candidate GLD-1-binding site (GBM 2) 300 base pairs (bp) downstream from GBM 1 in the ced-3 3′ UTR (Fig. 2A). A biochemical interaction between GLD-1 and ced-3 mRNA is also supported by the results of Scheckel et al. (2012), who performed GLD-1 immunoprecipitation with monoclonal antibodies followed by tiling array profiling of coprecipitated targets. In this data set, ced-3 mRNA is almost as highly enriched as the well-characterized GLD-1 target cep-1 (Fig. 2B).
Figure 2.
Two GBMs in the ced-3 3′ UTR mediate ced-3 mRNA repression by GLD-1. (A) GLD-1 binds to the ced-3 3′ UTR: HITS-CLIP of GLD-1::STREP/HA (Brümmer et al. 2013) and PAR-CLIP of GLD-1::GFP::Flag (Jungkamp et al. 2011) identified two candidate GBMs in the ced-3 3′ UTR. (B) Following GLD-1 immunoprecipitation, ced-3 mRNA showed a level of enrichment similar to that of the p53 tumor suppressor homolog cep-1, a known GLD-1 target (Schumacher et al. 2005). Data are from Scheckel et al. (2012). (C) The two GBMs are essential for GLD-1 binding to the ced-3 3′ UTR in vitro. Wild-type and GBM-mutated ced-3 3′ UTRs were biotinylated and tested for their ability to interact with GLD-1 from worm extracts. The constructs used are shown at the left. cep-1 was used as a positive control, and human rasm was used as a negative control. GLD-1's presence in the immunoprecipitation sample was verified by Western blot with monoclonal anti-GLD-1 antibodies. The right chart shows relative GLD-1 binding as measured by band intensity quantification obtained from four independent Western blot experiments. An asterisk represents the P-value for a paired t-test comparing the construct with wild-type ced-3 3′ UTR binding. (*) P < 0.005. (D) GBM mutations caused a strong increase of GFP::H2B expressions in the meiotic region of the germline. GFP images of adult gonads expressing GFP::H2B fused to either a wild-type ced-3 3′ UTR [Ppie-1::gfp::h2b::ced-3(3′ UTR)] or a ced-3 3′ UTR in which one (ced-3 GBM 1 mt 3′ UTR) or both (ced-3 GBM 1,2 mt 3′ UTR) GBMs were mutated. Dashed lines outline the gonads, and arrowheads indicate the positions of the distal tip cells. Bar, 20 µm. (E) Quantification of GFP::H2B fluorescence intensity in ced-3 3 3′ UTR reporters as a function of distance from the distal tip cell shows additive contribution of the two GBMs in ced-3 repression. Average fluorescence intensities of three animals are plotted. (F) Mean fluorescence intensity in the four distinct areas of the germline of ≥15 animals. Error bars represent SEM. n ≥ 15. See also Supplemental Figure S3.
To determine whether GLD-1 interacts with the GMB 1 and GMB 2 sites, we performed RNA affinity isolation experiments using synthetic biotinylated ced-3 transcripts added to worm lysates. While the wild-type ced-3 mRNA efficiently bound GLD-1, the GBM2 single mutant and the GBM1,2 double mutant significantly reduced GLD-1 binding (Fig. 2C).
We next verified the requirement of the two biochemically identified GBMs for translational repression of ced-3 mRNA by GLD-1 in vivo. To this end, we generated single-copy 3′ UTR reporter transgenic lines in which the expression of a GFP-tagged histone H2B protein (GFP::H2B) was under the control of either a wild-type ced-3 3′ UTR {opIs508[Ppie-1::gfp::h2b::ced-3(3′ UTR)]} or a ced-3 3′ UTR in which we introduced mutations in GBM 1 (ced-3 GBM 1 mt) or both GBMs (ced-3 GBM 1,2 mt). We observed a pronounced increase of the reporter expression in the early and late meiotic zones in the two reporters with mutated GBMs (Fig. 2D–F). The regions with increased ced-3 3′ UTR reporter expression coincided strongly with the regions where GLD-1 is expressed (Schumacher et al. 2005), suggesting that GLD-1 acts as a negative regulator on ced-3 mRNA. Moreover, the increase was additive, suggesting that both sites are important for ced-3 inhibition by GLD-1 in vivo. In order to exclude that the mutations in the two GBMs affected the binding of an unrelated RBP or of a microRNA (miRNA), we performed gld-1 RNAi in wild-type and ced-3 GBM 1,2 mt reporter strains. While the wild-type reporter was derepressed in the meiotic region of the germline, gld-1 RNAi did not cause any derepression of the ced-3 GBM 1,2 mt reporter (Supplemental Fig. S3). Taken together, our data suggest that GLD-1 inhibits ced-3 expression through direct binding to two GBMs in its 3′ UTR.
Post-transcriptional regulation of CED-3 is required for proper control of germ cell apoptosis
Having characterized the nature of GLD-1's interaction with ced-3, we moved on to the other candidates identified in the screen. The cgh-1(ok492) mutant recapitulated the RNAi screen results, causing an increase of CED-3::GFP translational reporter expression in the late meiotic and oocyte zones of the germline (Supplemental Fig. S4A). puf-8(ok302) similarly confirmed the RNAi results, showing an increase of CED-3::GFP expression in the mitotic region of the germline (Supplemental Fig. S4B).
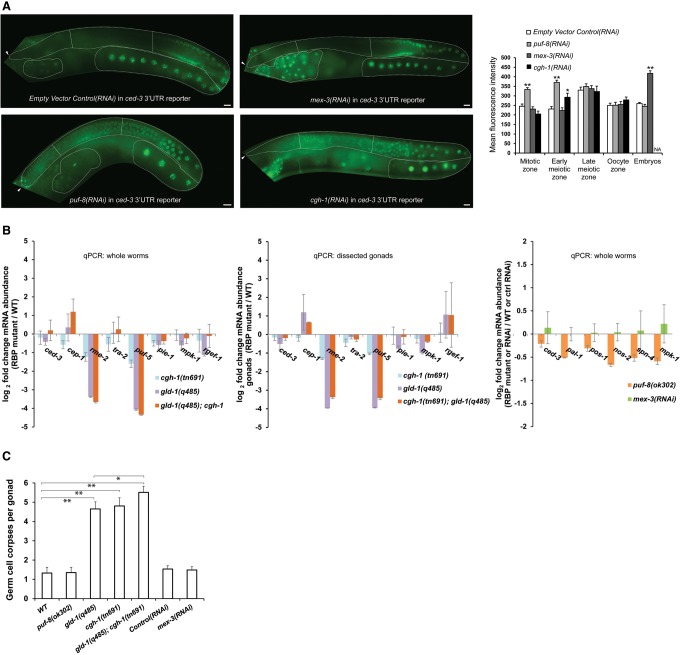
Furthermore, we investigated whether, similar to GLD-1, the RBPs PUF-8, MEX-3, and CGH-1 could interact with the ced-3 3′ UTR. puf-8(RNAi) and mex-3(RNAi) of transgenic animals expressing the ced-3 3′ UTR reporter {opIs508[Ppie-1::gfp::h2b::ced-3(3′ UTR)]} caused an increase in expression similar to that observed with the translational reporter (Fig. 3A). In contrast, cgh-1(RNAi) increased expression only in the early meiotic zone. This indicates that the ced-3 3′ UTR is sufficient for the effects exerted by PUF-8 and MEX-3, but not CGH-1, on its expression.
Figure 3.
Failure to translationally repress ced-3 in the death zone is associated with increased germ cell death apoptosis. (A) ced-3 3′ UTR is sufficient for exerting the PUF-8- and MEX-3-mediated repression of ced-3 expression. GFP images of adult gonads expressing the ced-3 3′ UTR reporter [Ppie-1::gfp::h2b::ced-3(3′ UTR)] treated with empty vector control(RNAi), puf-8(RNAi), mex-3(RNAi), and cgh-1(RNAi). Gonads are outlined with a dashed line, and arrowheads indicate the positions of the distal tip cells. Average mean fluorescence intensities measured in the four germline zones and fertilized embryos are plotted at the right. Different zones selected for quantification are separated with straight dashed white lines. Error bars represent SEM. n ≥ 10. Asterisks indicate significant P-values. (*) P < 0.05; (**) P < 0.00001. Bar, 10 µm. (B) gld-1(q485), cgh-1(tn691), gld-1(q485);cgh-1(tn691), puf-8(ok302), and mex-3(RNAi) animals show no changes in ced-3 mRNA abundance. (Charts 1,2) Quantitative RT–PCR (qRT–PCR) of known GLD-1 targets (cep-1, rme-2, tra-2, puf-5, pie-1, and mpk-1) (Wright et al. 2011) and a negative control rgef-1, a pan-neuronally expressed transcript in whole animals and gonads, respectively. As reported previously, GLD-1 and CGH-1 stabilize certain mRNA targets (rme-2 and puf-5) important for oocyte-to-embryo transition (Scheckel et al. 2012). (Chart 3) qRT–PCR of potential PUF-8 targets (pal-1, pos-1, spn-4, and mpk-1) and of the MEX-3 target nos-2 (Pagano et al. 2009; Mainpal et al. 2011; Pushpa et al. 2013) on whole animals. Results were normalized to a set of housekeeping and stably behaving transcripts (pgk-1, cdc-42, and Y45F10). mex-3(RNAi) data were compared with the empty vector control(RNAi). Error bars indicate SD from three (charts 1 and 2) and two (chart 3) biological replicates. (C) Physiological germ cell apoptosis is increased in gld-1(q485)-treated, cgh-1(tn691)-treated, and gld-1(q485);cgh-1(tn691)-treated but not in puf-8(ok302)-treated and mex-3(RNAi)-treated animals. Error bars represent SD from three independent experiments. n ≥ 15 animals per experiment. Asterisks indicate significant P-values from paired t-tests. (*) P < 0.05; (**) P < 0.0001.
Given the presence of four predicted PUF-8-binding motifs (PBMs) (Opperman et al. 2005) and two predicted MEX-3-binding motifs (MBMs) (Pagano et al. 2009) in the ced-3 3′ UTR (Supplemental Fig. S5A), we tested whether mutating those sites might derepress ced-3 3′ UTR expression. Mutations of all four PBMs and the two MBMs in the ced-3 3′ UTR did not alter the expression pattern of the reporter gene (Supplemental Fig. S5B,C), suggesting either the presence of additional binding sites or that PUF-8 and MEX-3 regulate ced-3 expression indirectly via other trans-acting factors.
Increased CED-3::GFP protein levels in gld-1, cgh-1, puf-8, and mex-3 RNAi could be a consequence of increased transcription, increased mRNA stability, or increased protein synthesis (or stability). To distinguish between these possibilities, we measured ced-3 mRNA levels in gld-1, cgh-1ts, gld-1; cgh-1ts, puf-8 mutants and mex-3(RNAi) animals. We detected no significant changes in ced-3 transcript levels compared with the wild-type and empty vector control(RNAi) controls, suggesting that ced-3 expression is repressed at the level of translation efficiency (or protein stability) (Fig. 3B).
In addition to RBPs, ced-3 expression might also be regulated by miRNAs, as has been reported for mammalian caspases (Wang et al. 2011, 2013; Fang et al. 2012). miRanda, a miRNA target prediction program (Betel et al. 2010), predicted the presence of three miRNA-binding sites (for miR-785, miR-86, and miR-792) in the ced-3 3′ UTR in close proximity to GBM 1. We wondered whether these miRNAs might synergize or compete with GLD-1 in regulating ced-3 expression in the germline. Scrambling the seed sequence of these three miRNA-binding sites had no effect on the ced-3 3′ UTR reporter expression (Supplemental Fig. S6), and crossing mir-86 and mir-785 deletion mutants into the CED-3::GFP reporter strain did not alter its germline expression. Moreover, the ced-3 transcript was not enriched following HITS-CLIP of the C. elegans Argonaute protein ALG-1 (Zisoulis et al. 2010), and no dedicated miRNA was identified in complex with the ced-3 transcript in ALG-1 iPAR-CLIP experiments, including ligation (Grosswendt et al. 2014), suggesting that C. elegans executioner caspase is not subject to extensive regulation by miRNAs.
We next questioned whether the increase in CED-3::GFP expression following loss of PUF-8, GLD-1, CGH-1, and MEX-3 is accompanied by an increase in apoptosis levels in the respective RBP mutants. Increased expression was correlated with higher germline apoptosis levels in gld-1 and cgh-1ts mutants but not puf-8 mutants and mex-3(RNAi) (Fig. 3C). Thus, increased CED-3 expression in the late meiotic region (death zone)—where cells normally die—leads to increased apoptosis, whereas CED-3 overexpression in other regions does not induce apoptosis, indicating that an increase in CED-3 expression alone is not sufficient to induce apoptosis and that additional factors are necessary for germ cell death.
Post-transcriptional regulation of human executioner caspase CASP3
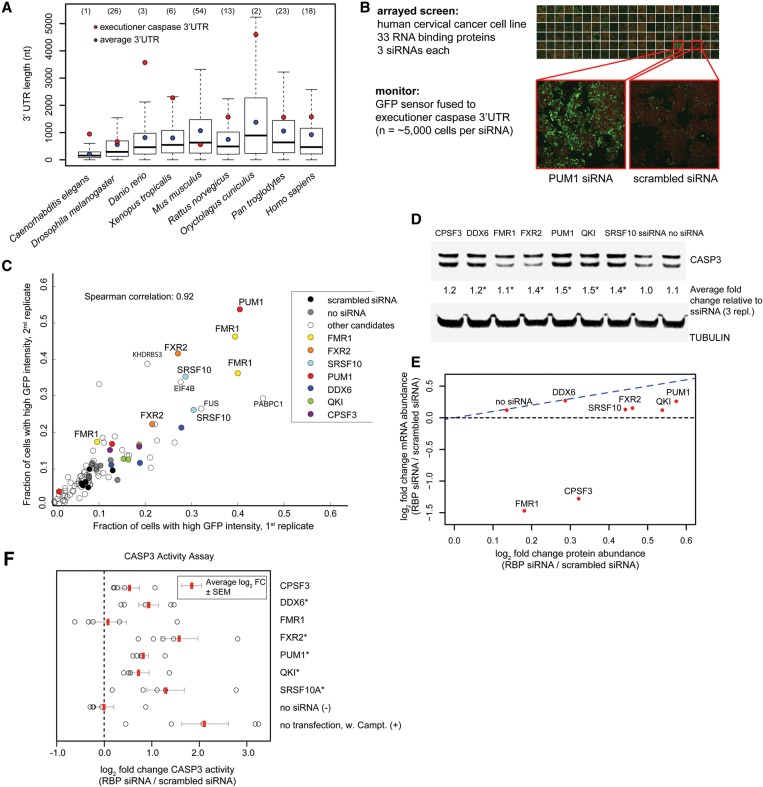
Is translational regulation of executioner caspase restricted to the C. elegans germline or could it be a more general phenomenon? C. elegans ced-3 has an unusually long 3′ UTR of 945 bp compared with the average 3′ UTR length in C. elegans of 213 bp (Supplemental Fig. S7A). The extensive length of the ced-3 3′ UTR made us explore the lengths of executioner caspase 3′ UTRs in other species. Strikingly, even though the average 3′ UTR length generally increases with organism complexity (Mazumder et al. 2003), we found that the 3′ UTR lengths of executioner caspases remained significantly larger than the average 3′ UTR in the majority of analyzed species (Fig. 4A; Supplemental Fig. S7). For example, caspase-3 3′ UTR lengths of zebrafish (Danio rerio), frogs (Xenopus tropicalis), and rabbits (Oryctolagus cuniculus) were in the top six percentile of all transcripts. While not proof, the conservation over long evolutionary times of an extremely long 3′ UTR does raise the possibility that regulation of apoptosis via translational control of caspase expression is likely not restricted to C. elegans but rather is a general strategy conserved across metazoans.
Figure 4.
Post-transcriptional regulation of human CASP3. (A) Executioner caspase 3′ UTRs are unusually long. Box plots show the distribution of 3′ UTR lengths across animal phyla. 3′ UTR lengths of executioner caspases (ced-3 in C. elegans, drICE in Drosophila, and caspase-3 in other species) are marked with a red dot. Median 3′ UTR length is represented with a line, average 3′ UTR is represented with a blue dot, and the whiskers extend to the most extreme data point, which is no more than 1.5 times the interquartile range from the box. Percentiles for the executioner caspase 3′ UTR length within a species are given in the parentheses above each organism. 3′ UTR lengths were extracted from the Ensemble Genes 75 database. (B) Image-based siRNA screen for post-transcriptional regulation of the caspase-3 3′ UTR. HeLa cells were treated with siRNAs against 33 RBPs. Caspase-3 levels were measured using the GFP::caspase-3 3′ UTR reporter (green), and cell outlines were detected by succinimidyl ester (red). (C) The fraction of cells with elevated level of GFP::caspase-3 3′ UTR reporter following knockdown of RBP. Three different siRNAs (represented as points) were used per gene. (D) Western blot of total CASP3 levels upon siRNA-mediated knockdown of candidate RBPs. The average fold changes in total CASP3 abundance following siRNA relative to scrambled siRNA and normalized to tubulin are shown. n = 3 biological replicates. (*) P < 0.05, paired t-test. (E) Comparison of mean caspase-3 protein with mRNA fold changes (log2) following knockdown of candidate RBPs. Caspase-3 protein and transcript levels (n = 3 biological replicates) were quantified by Western blot and bDNA single-molecule fluorescence in situ hybridization, respectively. (F) CASP3 activity following knockdown of the RBP candidates, measured via cleavage of the fluorogenic caspase-3 Ac-DEVD-AMC substrate. Treatment with the apoptosis-inducing agent camptothecin was used as a positive control. n = 5 biological replicates. Average log2 fold change (FC) in CASP3 activity (RBP siRNA/scrambled siRNA) is marked with red bar. Error bars represent SEM. (*) P < 0.05, paired t-test.
To experimentally test this hypothesis, we performed an image-based screen for translational regulators of caspase-3 in a human cervical cancer cell line (HeLa cells) (Fig. 4B). Thirty-three human candidate RBPs were selected for the screen based on either their homology with PUF-8, GLD-1, CGH-1, and MEX-3 or the presence of predicted or experimentally identified binding sites in the human caspase-3 3′ UTR by the RBPDB, CLIPz, and doRINA databases (Supplemental Table S2). We designed a GFP::caspase-3 3′ UTR reporter and measured its fluorescence intensity in single cells following cotransfection with three different siRNAs against the candidate genes. If a RBP were to regulate caspase-3 expression via binding to its 3′ UTR, knocking it down should result in a difference in reporter expression. We confirmed that the cell numbers between replicates were in good correlation (Spearman correlation 0.84) (Supplemental Fig. S8A), as were the GFP::caspase-3 3′ UTR reporter activities, irrespective of their specific readout (Supplemental Fig. S8B). For seven RBPs, at least two siRNAs resulted in a significant increase in reporter expression compared with the scrambled siRNA transfection (Fig. 4C; Supplemental Fig. S9). Three of these genes—PUM1, QKI, and DDX6—are homologs of C. elegans PUF-8, GLD-1, and CGH-1, respectively. The four additional candidates that negatively regulated the caspase-3 3′ UTR reporter were Fragile X mental retardation gene 1 (FMR1) and FXR2, serine/arginine-rich splicing factor 10 (SRSF10), and cleavage and polyadenylation-specific factor 3 (CPSF3). Interestingly, several of these RBPs, including PUM1, PUM2, FMR1, and FXR2, have been experimentally shown to bind to the caspase-3 3′ UTR (Galgano et al. 2008; Morris et al. 2008; Mukherjee et al. 2009; Hafner et al. 2010; Ascano et al. 2012). Moreover, the caspase-3 3′ UTR contains two sites that match the experimentally verified RNA-binding motif of QKI (Galarneau and Richard 2005). Several of the identified RBP-binding sites appear conserved between humans and closely related mammalian species (Supplemental Fig. S7F).
We next investigated whether the RBPs, in addition to the GFP sensor, may regulate endogenous CASP3 levels by knocking down the seven candidate RBPs and probing the total levels of CASP3 by a Western blot. Depletion of six out of seven candidates (all but CPSF3) resulted in a modest, but significant, increase of total CASP3 levels (Fig. 4D; Supplemental Fig. S10A), with knockdown of PUM1 and QKI causing the strongest (1.5-fold) up-regulation.
RBPs can influence gene expression at many levels, including mRNA processing, transport, localization, translation, and stability. To determine the effect of RBP knockdown on caspase-3 transcript level, we quantified caspase-3 mRNA in individual cells by bDNA single-molecule fluorescence in situ hybridization (FISH) (Battich et al. 2013). Knockdown of FMR1 and CPSF3 led to a twofold to threefold reduction in caspase-3 mRNA abundance, whereas RNAi of the other five RBPs led to very moderate increases in caspase-3 transcript levels (Supplemental Fig. S10B).
DDX6 showed very similar levels of mRNA and protein abundance changes, implying that the increase in caspase-3 protein levels could be largely explained by the corresponding increase in caspase-3 mRNA (Fig. 4E). In contrast, knockdown of the other six RBPs led to a greater increase in protein abundance than in mRNA abundance, suggesting that reduction in these RBPs increases CASP3 mRNA translation efficiency (or, alternatively, CASP3 protein stability). This effect was particularly striking for FMR1 and CPSF3, both of which showed mild increases in protein abundance despite the significant reduction in caspase-3 mRNA. GLD-1 was similarly found to both translationally repress and stabilize mRNAs of a certain portion of its targets (Scheckel et al. 2012). We conclude that mammalian CASP3, like C. elegans CED-3, can be regulated by RBPs at the post-transcriptional and likely translational levels.
Finally, we tested whether the increase in CASP3 levels is sufficient to alter the activity of CASP3 and thus possibly affect cell death levels in HeLa cells. To this end, we knocked down RBP expression by RNAi and measured CASP3 cleavage and enzymatic activity. Strikingly, depletion of five out of seven candidates identified in the screen (DDX6, PUM1, QKI, SRSF10, and FXR2) significantly increased CASP3 enzymatic activity (Fig. 4F). Knockdown of the C. elegans RBP homologs PUM1, QKI, and DDX-6 resulted in 1.8-fold, 1.6-fold, and 1.9-fold average increases of CASP3 cleavage, respectively. The strongest impact on CASP3 activity was observed following SRSF10 knockdown (2.5-fold average increase) and FXR2 knockdown (2.7-fold average increase). Our observations are consistent with the recent report indicating the role of SRSF10 in protecting DT40 cells from ER-induced stress apoptosis (Zhou et al. 2014). Given these results, we conclude that the large majority of hits obtained from the screen for post-transcriptional regulators of human caspase-3 indeed have an inhibitory effect on CASP3 expression and activity and thus, potentially, a physiological impact on apoptosis. Whether the RBPs identified here directly bind to and regulate the caspase-3 3′ UTR or act indirectly remains to be determined.
Discussion
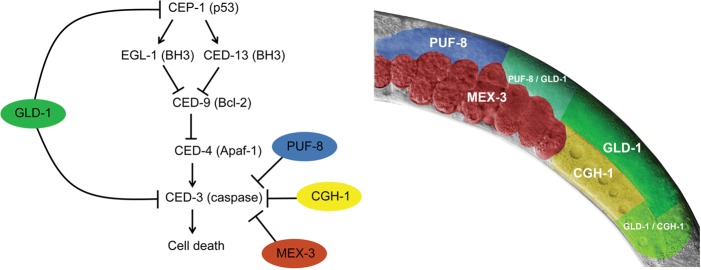
Here we show that four RBPs sequentially regulate executioner caspase expression in the C. elegans germline: PUF-8 in the mitotic and early meiotic zones, GLD-1 in the early and late meiotic zones, CGH-1 in the late meiotic zone, and oocytes and MEX-3 in embryos (Fig. 5). Regions where two RBPs coregulate CED-3 expression might be used to ensure a safe transition of repressed ced-3 mRNA between different germline zones without the hazard of unwanted cell death activation. We demonstrate that GLD-1 translationally represses ced-3 mRNA via two binding sites in its 3′ UTR, thereby enabling a dual control of cell death: upstream at the level of the p53 tumor suppressor cep-1 and at the level of the core apoptotic machinery via ced-3. Interestingly, the existence of a second proapoptotic target for GLD-1 had been predicted already by Schumacher et al. (2005), who noticed that loss of cep-1 function only partially suppressed the increase of germ cell apoptosis observed in gld-1(op236) mutants.
Figure 5.
Sequential regulation of ced-3 mRNA translation by PUF-8, GLD-1, CGH-1, and MEX-3. Model of sequential translation inhibition of ced-3 in the C. elegans germline and embryos. GLD-1 regulates apoptosis in two points of the apoptotic pathway: upstream at the level of p53 tumor suppressor cep-1 and at the point of no return, where it coregulates ced-3 together with PUF-8, CGH-1, and MEX-3. At the right is an image of the adult C. elegans gonad, adding spatial context to the ced-3 regulation by RBPs: PUF-8 inhibits ced-3 in the mitotic and early meiotic zones, GLD-1 inhibits ced-3 throughout the early and late meiotic zones, CGH-1 inhibits ced-3 in the late meiotic zone and oocytes, and MEX-3 inhibits ced-3 in the embryos. Whether MEX-3, PUF-8, and CGH-1 regulate ced-3 directly or indirectly via other factors remains to be determined.
Why do PUF-8 and MEX-3 regulate CED-3 expression in regions where CED-3 has no effect on apoptosis activation? One possibility would be that CED-3 has additional roles in the C. elegans germline. Indeed, nonapoptotic roles have been reported for caspases in both C. elegans and other species (Kuranaga and Miura 2007; Weaver et al. 2014).
The unusually long 3′ UTRs of executioner caspases across many species suggest that translational control might be an evolutionarily conserved mechanism for apoptotic regulation. Indeed, our screen in HeLa cells identified several conserved RBPs that influence expression and activation of human caspase-3. Three of these—PUM1, QKI, and DDX6—are homologs of the C. elegans RBPs that orchestrate CED-3 expression in the germline. The increase of total CASP3 levels following knockdown of these RBPs was generally of modest magnitude. However, as our results with C. elegans show, such changes can be of biological significance (particularly in sensitized backgrounds) when cells are on the verge of death. Moreover, given the significant number of regulators that we identified in our targeted screen, it appears likely that CASP3 can be regulated simultaneously by several RBPs and that the cumulative effect of the RBPs on total CASP3 protein levels will be significantly stronger.
Taken together, our results show that RBPs play an important role in the post-transcriptional regulation of CED-3 caspase in C. elegans—and possibly also of executioner caspases in other species, including humans. As both positive and negative dysregulation of apoptosis has been linked to various pathologies (Lessene et al. 2008; Schapira et al. 2014), modulation of RBP levels might provide a new therapeutic strategy to re-establish optimal apoptosis rates and thus treat disease.
Materials and methods
C. elegans strains
C. elegans strains were maintained at 20°C using standard procedures (Brenner 1974) unless indicated differently. Strains harboring cgh-1(tn691) were maintained at 15°C and shifted to 25°C at L4 stage for subsequent analysis of adult animals. Bristol strain N2 was used as the wild type. The following mutations and transgenes were used in this study: gld-1(q485)/ht2[qIs48], cgh-1(ok492)/ht2[qIs48], cgh-1(tn691), puf-8(ok302) unc-4(e120)/mIn1, opIs461[Pced-3::ced-3(genomic)::gfp::ced-3(3′ UTR)], opIs508[Ppie-1::gfp::h2b::ced-3(3′ UTR)], opIs507[Ppie-1::gfp::h2b::ced-3(3′ UTR) GBM 1 mt], opIs518[Ppie-1::gfp::h2b::ced-3(3′ UTR) GBM 1,2 mt], opIs520[Ppie-1::gfp::h2b::ced-3(3′ UTR) PBM 1,2,3,4 mt], opIs521[Ppie-1::gfp::h2b::ced-3(3′ UTR) MBM 1,2 mt], opIs516[Ppie-1::gfp::h2b::ced-3(3′ UTR) miR-785/86/792], and opIs517[Ppie-1::gfp::h2b::ced-3(3′ UTR) miR-785/86/792+GBM 1 mt].
RNAi and GFP quantification
RNAi was performed essentially as described (Neukomm et al. 2011). Animals were synchronized by bleaching and grown for 72 h after transfer to plates with bacteria expressing dsRNA against RBPs. The empty vector L4440 was used as a negative control. Transgenic animals expressing CED-3::GFP were anesthetized in a drop of 5 mM levamisole and inspected by differential interference contrast (DIC) microscopy and fluorescence microscopy for GFP. Mean fluorescence intensity from GFP photomicrographs of 10–30 adult hermaphrodites in four germline zones (mitotic, early meiotic, late meiotic, and oocyte) and embryos were quantified with ImageJ. Mean gray values measured in selected quadrant areas in respective germline and embryo zones were subtracted by the background intensity measured outside of the inspected animals. The mean fluorescence intensity of control reporters can vary between figures due to different exposure/gain settings used when capturing the images. Control and RNAi-treated/mutant animal images were always taken at the same time with same camera settings, allowing quantification comparisons between the two conditions. In Figure 2F, the gray value along a line starting from the distal tip cell, going through the mitotic and meiotic nuclei, and ending up in the oocyte was measured.
Transgenic lines
A low-copy CED-3::GFP translational reporter was generated via microparticle bombardment as described previously (Praitis et al. 2001). Phusion PCR was used to ligate ced-3 (genomic region) with the GFP on its C terminus. The fragment was then inserted into the pDONR 221 vector, which was subsequently recombined with pDONR P4-P1R containing the ced-3 promoter (5.6 kb upstream of ATG) and p2R-P3 containing the ced-3 3′ UTR (1.8 kb) according to the manufacturer's instructions (MultiSite Gateway, Life Technologies). Single-copy transgenes used for quantitative comparisons of the wild-type ced-3 3′ UTR and the ced-3 3′ UTR harboring mutations in RBPs and miRNA-binding sites were created by Mos1 transposase-mediated single-copy gene insertion (MosSCI) (Frøkjær-Jensen et al. 2012). Plasmids were generated in a Gateway LR reaction by recombining a short pie-1 promoter inserted into the pDONR P4-P1R vector (pCM1.127) (Merritt et al. 2008), gfp::h2b fusion in pDONR 221 (pBMF2.7) (Wright et al. 2011), and either wild-type or mutated versions of the ced-3 3′ UTR inserted into pDONR P2R-P3. Details on the mutations introduced in the ced-3 3′ UTR reporters via site-directed mutagenesis are in Supplemental Table S3. Primers used to introduce mutations are in Supplemental Table S4.
HITS-CLIP
The HITS-CLIP data set was generated as described previously (Brümmer et al. 2013). The data from the experiment are publicly available on the CLIPz server (http://www.clipz.unibas.ch).
Synthesis of biotinylated RNAs and RNA affinity isolation experiments
All DNA templates for biotin RNA synthesis were prepared by PCR from genomic DNA obtained from wild-type worms with 5′ oligonucleotides bearing a T7 RNA polymerase promoter sequence. Oligonucleotide primers are in Supplemental Table S4. Biotinylated RNAs were produced with T7-RNA polymerase and biotin RNA labeling mix (Roche, 11685597910) as instructed by the manufacturer. Biotin RNA pull-down experiments were performed essentially as described (Gerber et al. 2006). Extracts were prepared by mechanical disruption (100 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM DTT, 0.75% IGEPAL, 0.1 mg/mL heparin, 0.1 mg/mL tRNA, complete EDTA-free protease inhibitor cocktail [Roche, 11836170001]) from synchronized L4 worms. Two-hundred micrograms (protein content) of extract was incubated with 5 pmol of biotinylated RNAs. RNA–protein complexes were further captured with streptavidin beads (Invitrogen, 11205D), resolved on 10% SDS–polyacrylamide gels, and transferred to PVDF membranes (Thermo Scientific) for immunoblot analysis. Therefore, membranes were blocked in 5% low-fat milk dissolved in phosphate-buffered saline/0.1% Tween-20 (PBST) for 1 h at room temperature. Membranes were probed overnight at 4°C with mouse anti-GLD-1 (1:30) (Scheckel et al. 2012) in 2% low-fat milk in PBST and finally probed with HRP-linked anti-mouse (1:2500) for 1 h at room temperature. Membranes were developed with the enhanced chemiluminescence detection kit (Amersham).
Quantitative RT–PCR (qRT–PCR)
Around 200 synchronized young adults that had very few to no eggs from wild-type (12 h after L4), gld-1(q485), cgh-1(tn691), gld-1(q485);cgh-1(tn691), puf-8(ok302), empty vector control(RNAi), and mex-3(RNAi) conditions were collected for relative quantification of transcripts. Total RNA was isolated with Trizol (Life Technologies) according to the manufacturer's instructions and reverse-transcribed with cDNA synthesis kit (SuperScript III Platinum, Invitogen). The forward and reverse oligos used for qPCR are in Supplemental Table S4. pgk-1, cdc-42, and Y45F10.4 were used to normalize transcript levels.
For RNA extraction from gonads, gonads of 50 animals staged as above were dissected and lysed in PicoPure RNA isolation kit (Roche) buffer according to the manufacturer's protocol.
Germ cell apoptosis
Bleach-synchronized animals were grown at 15°C and shifted to 25°C at L4/adult molt. Germ cell corpses were scored 32 h after L4/adult molt in all strains except gld-1(q485);cgh-1(tn691), which was developmentally delayed (needed 25% more time to reach the L4/adult molt stage than wild type) and scored 43 h after L4/adult molt. Worms were anesthetized in a drop of 5 mM levamisole, and germ cell corpses were counted using DIC microscopy. For empty vector control and mex-3 RNAi experiments, synchronized L1 animals were transferred to the NGM agarose plates with bacteria expressing the respective RNAi clone as described above.
Cell culture
HeLa Kyoto cells were kindly provided by J. Ellenberg (EMBL). Cells were tested for identity by karyotyping and tested for the absence of mycoplasma before use. Culturing was done in DMEM (Gibco) supplemented with 10% FCS and 1% glutamine (Gibco) and, during experiments, additionally supplemented with 1% penicillin/streptomycin (Gibco). Cells were incubated at 37°C, 95% humidity, and 5% CO2.
siRNA screen on the human caspase-3 3′ UTR reporter
Transfection mixes were prepared by mixing equal volumes of 0.5 µM Silencer Select siRNAs (Ambion) in OptiMem (Life Technologies) and 1% Lipofectamine 2000 (Life Technologies) in OptiMem. For each gene, three different siRNAs were reverse-transfected in two wells of a plastic-bottomed µClear 384-well plate (Greiner): In each well, 700 HeLa Kyoto cells suspended in 40 µL of medium were added on top of 10 µL of transfection mix. After 3 d of culturing, each well was forward-transfected with 100 ng of pDEST53-GFP::caspase-3 3′ UTR and 0.5% Lipofectamine 2000 according to the manufacturer's protocol. After 37 h, cells were washed with PBS, fixed for 30 min in 4% EM-grade paraformaldehyde (EMS) in PBS, permeabilized for 10 min in 0.1% Triton X-100 in PBS, stained for 10 min with 0.2 µg/mL 4,6-diamidino-2-phenylindole in PBS, and incubated for 5 min in 1 ng/µL Alexa fluor 647 carboxylic acid and succinimidyl ester (Invitrogen) in carbonate buffer (1.95 mL of 0.5 M NaHCO3, 50 µL of 0.5 M Na2CO3, 8 mL of water) to mark the outline of single cells. Images were acquired on an automated spinning-disk microscope from Yokogawa (CellVoyager 7000) with an enhanced CSU-X1 spinning disk (Microlens-enhanced dual Nipkow disk confocal scanner, wide view type), a 40× Olympus objective of 0.95 NA, and Neo sCMOS cameras (2560 × 2160 pixels; Andor). Computational object identification, illumination correction, background subtraction, and quality control were performed as described previously (Stoeger et al. 2015). For each well, the activity of the reporter was quantified by measuring the fraction of cells for which the median GFP intensity within the nucleus exceeded 10 grayscale values.
Caspase activity reporter
HeLa Kyoto cells were reverse-transfected with one siRNA per gene as described above. Cells were washed at 37°C with 1× caspase cleavage buffer (0.1 M HEPES at pH 7.5, 10% sucrose, 0.1% CHAPS) and incubated with 37°C Ac-DEVD-AFC reaction mix (1× caspase cleavage buffer, 10 mM DTT, 333 µM Ac-DEVD-AFC [Enzo Life Sciences]) containing 7.5 µM DRAQ5 (Thermo Scientific). Samples were incubated at 37°C within a CellVoyager 7000 microscope and imaged after 55 min using a 10× objective. Single cells were segmented using DRAQ5, and reporter activity was determined by averaging the mean blue intensities of every single cell.
Western blot
Extracts from HeLa Kyoto cells were prepared from cells reverse-transfected with siRNA as described above. One siRNA per gene was selected based on the strongest phenotype in the screen and the lowest article number (indicative of the best possible design). Apoptosis was induced by treating the cells with 25 µM camptothecin in DMEM (10% FCS, 1% glutamine) for 13 h. Around 500,000 cells were detached from wells with lysis buffer (150 mM NaCl, 50 mM HEPES at pH 7.5, 1% Triton, 0.1% SDS, 2 mM DTT, 5 mM EDTA, protease inhibitor tablet) followed by two rounds of sonication (15-sec sonication + 15-min break on ice). The concentration of cleared protein extracts was determined using a Protein 660-nm assay (Pierce). Samples were heated in NuPage buffer (Life Technologies) and loaded onto a 4%–12% Bis-Tris NuPage gel. After SDS-PAGE, the gel was transferred to a nitrocellulose membrane using a wet blot chamber (Invitogen, X cell II blot). The membranes were blocked with 5% milk in TBST for 1 h followed by probing with caspase-3 antibody (1:2000; Cell Signalling, 9662) and tubulin antibody (1:5000; Sigma, T6074) as instructed by the vendors.
bDNA single-molecule FISH
Caspase-3 transcript molecules were detected as fluorescent spots by bDNA single-molecule FISH using ViewRNA reagents (Affymetrix) on an automated experimental platform as described before (Battich et al. 2013) except that the optional protease treatment was omitted. Total protein of single cells was measured by Alexa fluor 647 carboxylic acid and succinimidyl ester (Invitrogen), and spot counts were normalized by division with total protein.
Supplementary Material
Acknowledgments
We thank Martin Moser for technical support with qRT–PCR; Michael Daube for help in the generation of transgenic strains; Rafal Ciosk for providing the gld-1;cgh-1 double mutant, GFP:H2B vector, GLD-1 monoclonal antibodies, and input; Lynn Wong for critical input; Jan Ellenberg for providing HeLa Kyoto cells; and members of the Hengartner laboratory for critical discussions on the project. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was funded by a Swiss National Science Foundation Sinergia grant (CRSII3_141942). D.S. and S.E. performed C. elegans experiments (RNAi screen and ced-3 3′ UTR reporter transgene generation and image analysis). T.S. performed the siRNA screen in HeLa cells, caspase activity assays, and the required computational analyses. D.S. performed the qRT–PCR experiments, apoptosis assays, and Western blots and analyzed the 3′ UTRome of different species. A.M.M.-G. performed GLD-1 RNA affinity isolations and analyzed the presence of RBP sites in human caspase-3. L.X. generated the CED-3::GFP transgenic line. J.I., X.Z, P.G., R.E., and R.H. helped with experimental design and procedures. D.S., T.S., A.M.M.-G., A.P.G., L.P., and M.O.H. designed the experiments. D.S. and M.O.H. wrote the manuscript. All authors provided detailed comments.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.285726.116.
Freely available online through the Genes & Development Open Access option.
References
- Ariz M, Mainpal R, Subramaniam K. 2009. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev Biol 326: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A, Gartner A. 2013. Germ cell apoptosis and DNA damage responses. Adv Exp Med Biol 757: 249–276. [DOI] [PubMed] [Google Scholar]
- Battich N, Stoeger T, Pelkmans L. 2013. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods 10: 1127–1133. [DOI] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag PR, Nakamura A, Blackwell TK. 2005. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development 132: 4975–4986. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer A, Kishore S, Subasic D, Hengartner M, Zavolan M. 2013. Modeling the binding specificity of the RNA-binding protein GLD-1 suggests a function of coding region-located sites in translational repression. RNA 19: 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Lambie EJ, Bindu S, Mikeladze-Dvali T, Conradt B. 2015. Engulfment pathways promote programmed cell death by enhancing the unequal segregation of apoptotic potential. Nat Commun 6: 10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R. 2006. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311: 851–853. [DOI] [PubMed] [Google Scholar]
- Conradt B. 2009. Genetic control of programmed cell death during animal development. Annu Rev Genet 43: 493–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. 1996. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87: 205–216. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH. 1999. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 68: 383–424. [DOI] [PubMed] [Google Scholar]
- Fang J, Song X, Tian J, Chen H, Li D, Wang J, Ren A, Yuan W, Lin L. 2012. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis 17: 410–423. [DOI] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T. 1995. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. 2011. Programmed cell death in animal development and disease. Cell 147: 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A, Richard S. 2005. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol 12: 691–698. [DOI] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. 2008. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One 3: e3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK. 2008. Germline survival and apoptosis. In WormBook (ed. The C. elegans Research Community), WormBook, doi: 10.1895/wormbook.1.145.1; http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Shi Y, Nakagawa A, Yoshina S, Mitani S, Shi Y, Xue D. 2008. Inhibition of CED-3 zymogen activation and apoptosis in Caenorhabditis elegans by caspase homolog CSP-3. Nat Struct Mol Biol 15: 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Zhou QH, Kage-Nakadai E, Shi Y, Yan N, Mitani S, Xue D. 2009. Caenorhabditis elegans caspase homolog CSP-2 inhibits CED-3 autoactivation and apoptosis in germ cells. Cell Death Differ 16: 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. 2014. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell 54: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P. 2010. IAPs: from caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat Rev Cancer 10: 561–574. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol 180: 165–183. [DOI] [PubMed] [Google Scholar]
- Jungkamp A, Stoeckius M, Mecenas D, Grün D, Mastrobuoni G, Kempa S, Rajewsky N. 2011. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell 44: 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritikou EA, Milstein S, Vidalain P, Lettre G, Bogan E, Doukoumetzidis K, Gray P, Chappell TG, Vidal M, Hengartner MO. 2006. C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival. Genes Dev 20: 2279–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E, Miura M. 2007. Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol 17: 135–144. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. 2007. Caspases in cell survival, proliferation and differentiation. Cell Death Differ 14: 44–55. [DOI] [PubMed] [Google Scholar]
- Lee MH, Schedl T. 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev 15: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessene G, Czabotar PE, Colman PM. 2008. BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov 7: 989–1000. [DOI] [PubMed] [Google Scholar]
- Lettre G, Kritikou EA, Jaeggi M, Calixto A, Fraser AG, Kamath RS, Ahringer J, Hengartner MO. 2004. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ 11: 1198–1203. [DOI] [PubMed] [Google Scholar]
- Mainpal R, Priti A, Subramaniam K. 2011. PUF-8 suppresses the somatic transcription factor PAL-1 expression in C. elegans germline stem cells. Dev Biol 360: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer CW, Chiorazzi M, Shaham S. 2007. Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development 134: 1357–1368. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. 2003. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci 28: 91–98. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18: 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. 2008. Ribonomic analysis of human Pum1 reveals cis–trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol 28: 4093–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. 2009. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol 5: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK. 2001. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development 128: 3221–3232. [DOI] [PubMed] [Google Scholar]
- Neukomm LJ, Frei AP, Cabello J, Kinchen JM, Zaidel-Bar R, Ma Z, Haney LB, Hardin J, Ravichandran KS, Moreno S, et al. 2011. Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat Cell Biol 13: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. 2005. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol 12: 945–951. [DOI] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, Essien KI, Ryder SP. 2009. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc Natl Acad Sci 106: 20252–20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpa K, Kumar GA, Subramaniam K. 2013. PUF-8 and TCER-1 are essential for normal levels of multiple mRNAs in the C. elegans germline. Development 140: 1312–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racher H, Hansen D. 2012. PUF-8, a Pumilio homolog, inhibits the proliferative fate in the Caenorhabditis elegans germline. G3 (Bethesda) 2: 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, Olanow CW, Greenamyre JT, Bezard E. 2014. Slowing of neurodegeneration in Parkinson's disease and Huntington's disease: future therapeutic perspectives. Lancet 384: 545–555. [DOI] [PubMed] [Google Scholar]
- Scheckel C, Gaidatzis D, Wright JE, Ciosk R. 2012. Genome-wide analysis of GLD-1-mediated mRNA regulation suggests a role in mRNA storage. PLoS Genet 8: e1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee M, Nayak S, Volkmann K, Hofmann ER, Hofmann R, Hengartner M, Schedl T, Gartner A. 2005. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. [DOI] [PubMed] [Google Scholar]
- Stoeger T, Battich N, Herrmann MD, Yakimovich Y, Pelkmans L. 2015. Computer vision for image-based transcriptomics. Methods 85: 44–53. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. 2003. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the Pumilio-like protein PUF-8. Curr Biol 13: 134–139. [DOI] [PubMed] [Google Scholar]
- Tomazella GG, Kassahun H, Nilsen H, Thiede B. 2012. Quantitative proteome analysis reveals RNA processing factors as modulators of ionizing radiation-induced apoptosis in the C. elegans germline. J. Proteome Res 11: 4277–4288. [DOI] [PubMed] [Google Scholar]
- Wang H, Yu X, Liu Z, Cheng X, Samartzis D, Jia L, Wu S, Huang J, Chen J, Luo Z. 2011. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J. Pathol 225: 232–242. [DOI] [PubMed] [Google Scholar]
- Wang K, Liu F, Zhou L, Ding S, Long B, Liu C, Sun T, Fan Y, Sun L, Li P. 2013. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis 4: e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BP, Zabinsky R, Weaver YM, Lee ES, Xue D, Han M. 2014. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. Elife 3: e04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R. 2011. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J 30: 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wu W, Li H, Cheng Y, Wei N, Zong J, Feng X, Xie Z, Chen D, Manley JL, et al. 2014. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res 42: 4019–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. 2010. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.