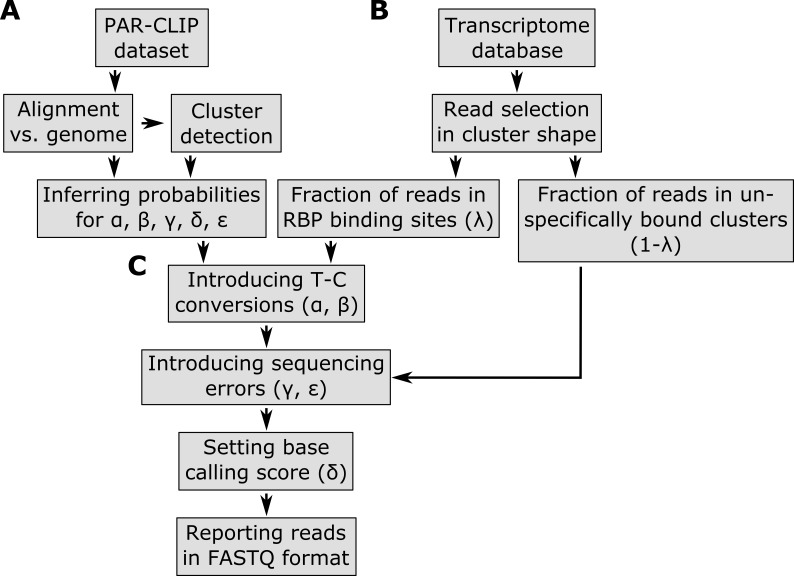
Figure 1. Pipeline of the PAR-CLIP read simulator implemented in the PARA-suite.
Part A describes the process of generating the error profile and other parameters learned from a real PAR-CLIP dataset. Part B starts to generate reads mapping to RBP binding sites (clusters) on transcript regions from a given transcript database (e.g., Ensembl genes). In Part C, the pre-calculated profiles are used to introduce T–C conversions, sequencing errors, indels and base-calling quality scores to the defined reads.