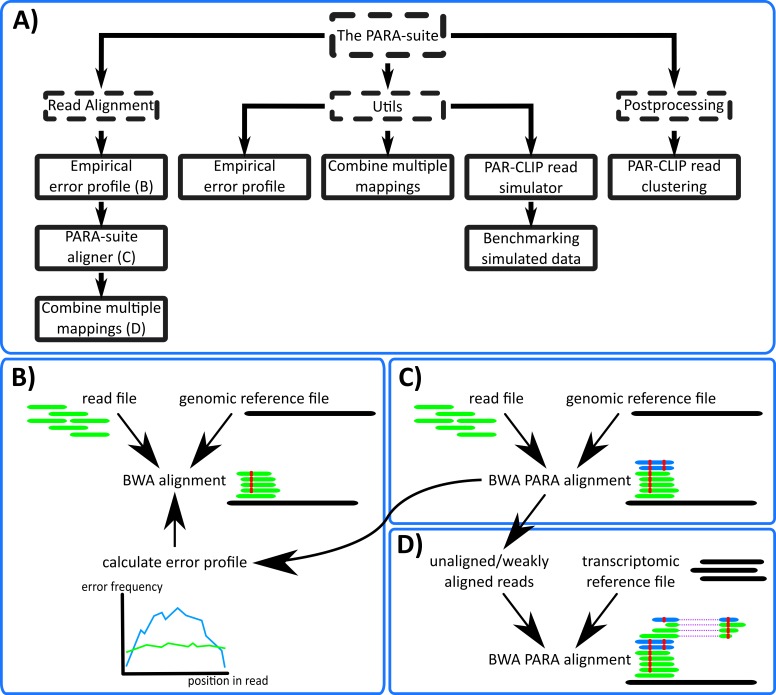
Figure 2. The PARA-suite.
(A) The PARA-suite. Dashed boxes represent software packages; all other boxes represent executable programs. The Utils package includes tools for working with error-prone sequencing data and the postprocessing package contains a tool for clustering an aligned PAR-CLIP dataset to identify RBP-bound genomic regions. (B) Read alignment by a fast read aligner is necessary to infer the error profile for a particular read dataset (we selected BWA). (C) BWA PARA is applied to the entire dataset to map error-prone reads, indicated here by the additional mapping of the two reads (shown in blue). (D) An optional alignment versus a transcriptome reference database can be executed using BWA PARA to identify previously unmapped reads.