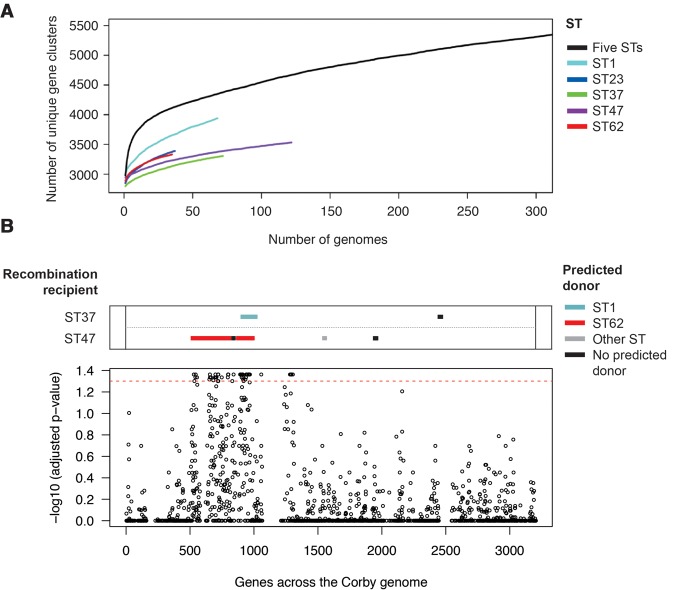
Figure 5.
Gene content analysis and the nucleotide diversity of core genes within the five disease-associated STs. (A) Rarefaction curves applied to the strains of L. pneumophila ST1 (71 isolates), ST23 (37 isolates), ST37 (72 isolates), ST47 (122 isolates), ST62 (35 isolates), and all five STs together (337 isolates), showing that differences in gene content exist among the five STs, but that the number of novel genes in the overall pan-genome is beginning to plateau. (B) Log-transformed P-values derived from testing whether the five disease-associated STs have lower than expected nucleotide diversity values in individual core genes given their nucleotide diversity across all 1888 core genes, and with respect to the gene conservation across the species (excluding isolates from the distant subspecies, strains ST5 and ST152, which are nested within ST1, and strains ST36 [Philadelphia], ST42, and ST578 [Alcoy], which are also disease-associated strains). The core genes are ordered as in the Corby genome. Any noncore genes (genes in <100% isolates) are omitted. The horizontal dotted red line indicates the significance threshold when the Benjamini-Hochberg method is applied to correct for multiple testing. The box at the top shows the location and predicted origins of recombined regions that were detected on the branches leading to the ST37 and ST47 lineages. Recombined regions that were found in the ST37 and ST47 accessory genomes are not shown.