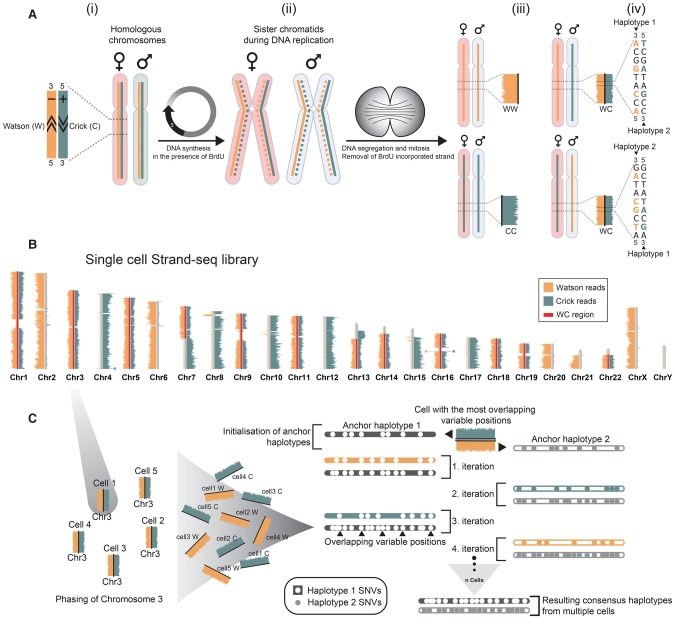
Figure 1.
Direct whole-chromosome haplotyping using single-cell template strand sequencing (Strand-seq). (A,i) Two homologous chromosomes, one originating from the mother (light red) and one from the father (light blue), are shown. Each homolog is composed of a positive template strand (Crick; teal) and a negative template strand (Watson; orange). (ii) Cells incorporate BrdU during DNA replication, generating hemi-substituted sister chromatids containing one BrdU-negative template strand (solid line) and one BrdU-positive newly synthesized strand (dashed line). (iii) Segregation of sister chromatids in two daughter cells follows the depicted combinations of maternal and paternal template strands. The newly formed DNA strands containing BrdU are selectively removed in daughter cells during library preparation, such that only the original template DNA strands are sequenced. Read density along a chromosome is plotted as horizontal bars. (iv) When daughter cells inherit one Crick and one Watson template strand for a particular chromosome, we can use strand directionality to directly assign all reads to separate haplotypes. (B) Example of a single-cell Strand-seq library, generated from HapMap cell line NA12878. Each chromosome is represented as a vertical ideogram, and the distribution of directional sequencing reads is represented as horizontal lines along each chromosome, with Watson in orange and Crick in teal. WC regions that were selected for haplotype phasing are highlighted by red bars. (C) The custom phasing algorithm StrandPhase processes one chromosome at a time. Cells that inherit one Crick and one Watson template strand for a particular chromosome are selected as input, and the SNVs identified on each template strand are used to derive each single-cell haplotype. In the first iteration, anchor haplotypes are established by pairing single-cell haplotypes exhibiting the highest number of overlapping heterozygous SNVs. This is used to initialize the consensus haplotypes “H1” and “H2,” which are further built upon in subsequent iterations. In the second iteration, the second most-dense single-cell haplotype is considered and compared to both consensus haplotypes, and any new SNVs are added to the consensus haplotype showing the best concordance. With each iteration, the consensus haplotypes are extended until no additional single-cell haplotype can be reliably assigned to the one of the consensus haplotypes.