Figure 2.
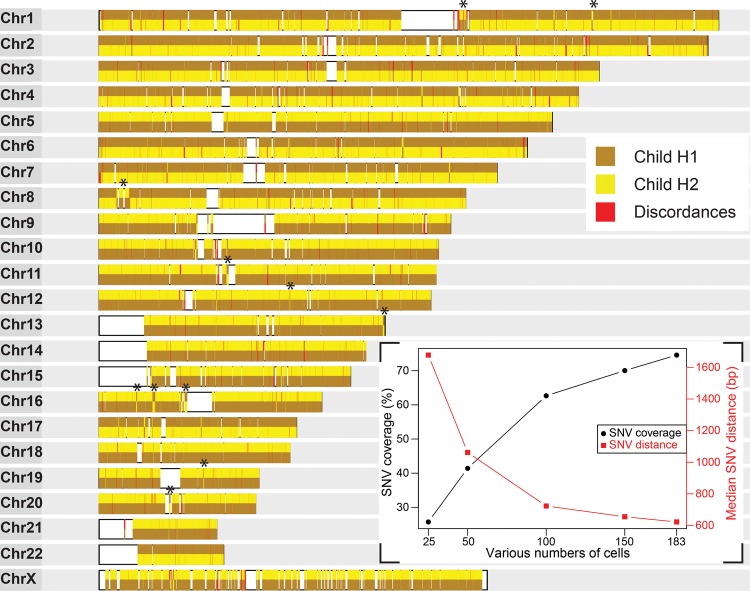
Accurate and dense whole-genome haplotypes are built from multiple single-cell Strand-seq libraries. Assembled haplotypes of the child derived from 183 Strand-seq libraries. Chromosome ideograms illustrate 151,700 high-confidence (covered in more than one cell) heterozygous SNV positions phased from Strand-seq data and compared with the HapMap reference. The consensus haplotypes determined by Strand-seq are depicted for each chromosome, with each SNV represented by a vertical line and color-coded based on whether it matched the child's reference homolog 1 (brown) or homolog 2 (yellow) listed in the HapMap reference. The contiguous haplotypes extend the whole length of each chromosome, spanning centromeres and reference assembly gaps (white blocks). Discordant alleles that did not match either reference haplotype are shown in red. (Asterisks) Short localized switches in haplotypes that were confirmed as homozygous inversions. (Inset) The percentage of HapMap reference SNVs covered (black line) and the median distance between these SNVs (red line) are plotted for various numbers of single-cell libraries (25, 50, 100, 150), randomly sampled from the entire data set of 183 cells.