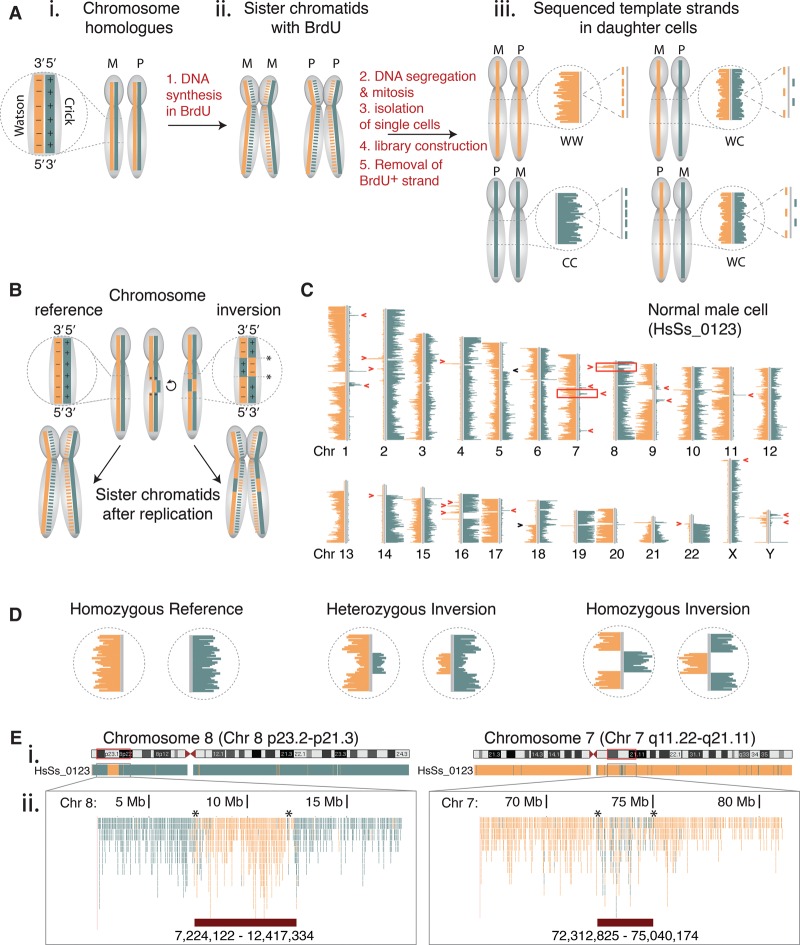
Figure 1.
Visualizing inversions at high resolution in single cells by template strand sequencing (Strand-seq). (A) Inheritance patterns of template strands during mitotic division. (i) Maternal (M) and paternal (P) chromosome homologs consist of complementary but unique DNA strands, called Watson (W, minus strand, orange solid line) and Crick (C, plus strand, teal solid line). (ii) DNA synthesis incorporates BrdU into newly formed strands (dotted lines). These are selectively removed during Strand-seq library construction to generate sequencing reads from the template strands only. (iii) When aligned to the reference genome and represented on chromosomal ideograms (using BAIT software), the possible combinations of template strand inheritance for each homolog in daughter cells is shown. For a diploid cell, the template strands of any given chromosome can be inherited as WW, CC, or WC, where each strand represents either the maternal or paternal homolog. (B) Inversions are a localized reorientation in the W-C state along each single DNA strand of a chromosome, with respect to the reference genome. Asterisks denote breakpoints. (C) A single Strand-seq library from an adult male shows the template strand inheritance patterns of all chromosomes (Chr). Inversions appear as segmental changes in strand orientation along a chromosome (red arrowheads) and are distinguished from sister chromatid exchange events (black arrowheads). Boxed regions on Chr 7 and Chr 8 are previously described inversions. (D) Expected Strand-seq results for each possible inversion genotype. (E) UCSC Genome Browser view of Strand-seq data of cell shown in C, BED-formatted and uploaded as custom tracks for Chr 8p23 (homozygous inversion, left panel) and Chr 7q11 (heterozygous inversion, right panel). (i) Whole chromosome ‘packed’ view of aligned Crick (teal) and Watson (orange) reads. (ii) Zoom of genomic regions containing inversions denoted by boxed regions in C. In this ‘squished’ view, each single aligned C and W read is denoted as an individual teal or orange line, and the chromosomal coordinates of the inversions (lower red bars) are manually mapped as the last base pair position of the 5′ and 3′ read flanking the inversion (asterisks).