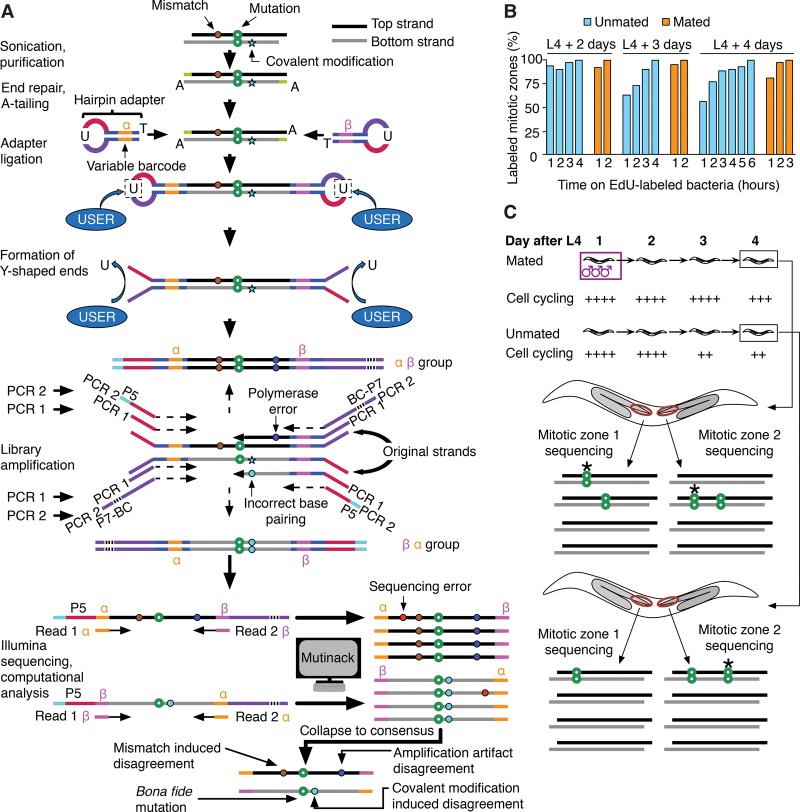
Figure 1.
Preparation of sequencing libraries, de novo mutation calling, and overall experimental scheme of SIP-HAVA-seq. (A) DNA to sequence is sheared, end-repaired, and, before any amplification is performed, ligated to hairpin adapters comprising a variable barcode. After linearization using NEB's “USER” enzyme, a first PCR is performed to amplify the material, and a second to add Illumina sequencing adapters. Illumina reads that correspond to the same “duplex” are grouped using the custom program “Mutinack” that relies on mapping position and variable barcode sequence. Mutation candidates are only reported if they are supported by both top and bottom strand amplification products. Mutinack also reports disagreements between the top and bottom strands, which can be indicative of a mismatch or damage-induced covalent modification in the original piece of DNA (since covalent modification can cause incorrect base-pairing during replication) or of an in vitro amplification error that occurred at an early step of the PCR; this step of the analysis does not distinguish between the two potential sources of disagreement. (BC) Illumina multiplexing barcode; (P5, P7) flow cell attachment sites. (B) EdU continuous labeling time course in hermaphrodites, taken at ages spanning the interval between onset of adulthood and time of collection for sequencing. (C) Experimental scheme. Each library is created from a single dissected worm germline MZ (red oval), comprising ∼250 cells. Mutations detected in one MZ but not the sister MZ from the same worm must have arisen de novo (stars in diagram). The speed of MZ cell cycling is increased by mating, and one can thus compare mutation rates in germ cells cycling at different speeds.