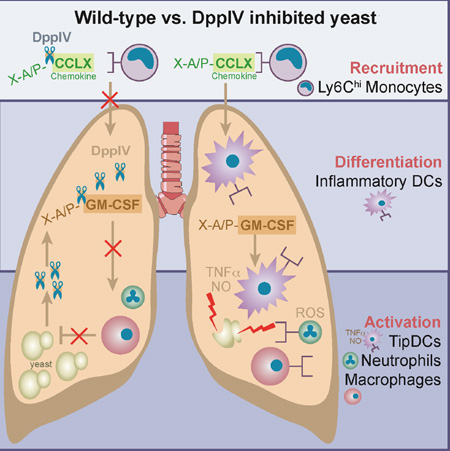
Abstract
Systemic fungal infections trigger marked immune-regulatory disturbances, but the mechanisms are poorly understood. We report that the pathogenic yeast of Blastomyces dermatitidis elaborate dipeptidylpeptidase IVA (DppIVA), a close mimic of the mammalian ectopeptidase CD26, which modulates critical aspects of hematopoiesis. We show that, like the mammalian enzyme, fungal DppIVA cleaved C-C chemokines and GM-CSF. Yeast producing DppIVA crippled the recruitment and differentiation of monocytes, and prevented phagocyte activation and ROS production. Silencing fungal DppIVA gene expression curtailed virulence and restored recruitment of CCR2+ monocytes, generation of TipDC, and phagocyte killing of yeast. Pharmacological blockade of DppIVA restored leukocyte effector functions and stemmed infection, while addition of recombinant DppIVA to gene-silenced yeast enabled them to evade leukocyte defense. Thus, fungal DppIVA mediates immune-regulatory disturbances that underlie invasive fungal disease. These findings reveal of form of molecular piracy by a broadly conserved aminopeptidase during disease pathogenesis.
Keywords: Fungal, TipDCs, macrophages, neutrophils, DppIV
Graphical abstract
eTOC BLURB
Mammalian proteases regulate the immune response by altering the activity of cytokines and chemokines. Here, Sterkel, Lorenzini and colleagues identify a fungal protease that promotes virulence and suppresses innate immunity to infection by mimicking the immune-modulatory activity of its mammalian counterpart.
INTRODUCTION
Pathogenic fungi have been dubbed the hidden killers due to the mounting rates of fungal infections. Immune compromised patients such as those with AIDS, organ transplants, and cancer and chemotherapy are among those at risk of serious fungal infections. The endemic dimorphic fungi are primary pathogens that collectively account for nearly one million systemic infections annually in North America (Pfaller and Diekema, 2010). These agents infect previously healthy individuals, but can also reactivate from a latent state when immunity is impaired. Among these systemic mycoses, the proportion of immune competent persons with symptomatic illness varies according to pathogen. At one end of the spectrum, about 50% become overtly ill after infection with Blastomyces dermatitidis (Klein et al., 1986), whereas about 10% manifest clinically significant illness with Histoplasma capsulatum (Ward et al., 1979). The high ratio of illness to infection and the potential severity of disease underscore the pathogenic and immune evasive potential of dimorphic fungi and make them challenging pathogens from a clinical vantage point.
Several factors have been linked to virulence in dimorphic fungi (Rappleye and Goldman, 2008). Some include calcium binding protein (CBP) and superoxide dismutase (SOD) in H. capsulatum, glucan synthase in Coccidiodes sp., α-1,3-glucan and Drk1 (dimorphism regulating kinase) in several members, and Blastomyces adhesin-1 (BAD-1) in B. dermatitidis. Immune dysregulation is a hallmark of infections with dimorphic fungi, but there is limited insight about how fungal factors subvert immunity. SOD protects Histoplasma from oxidative stress (Youseff et al., 2012), and surface α-1,3-glucan shields this fungus from recognition by dectin-1 (Rappleye et al., 2007). BAD-1 has multiple functions: it mediates binding of Blastomyces to macrophages (Mϕs) and lung tissue; modulates expression of host TNF-α and TGF-β; binds calcium and other divalent cations; and impairs T cell activation and function by engaging heparin sulfate modifications of surface CD47 (Brandhorst et al., 2013).
Failure of vaccination at the lung mucosa reveals features of immune dysregulation induced by dimorphic fungi. An attenuated, BAD-1 deletion strain of B. dermatitidis, which protects against lethal experimental blastomycosis when given subcutaneously, fails to protect when given via the respiratory route (Wuthrich et al., 2012). Vaccine delivery at the respiratory mucosa induces a host immune regulatory circuit, which hampers the recruitment of Ly6Chi monocytes into the lungs, and undermines the priming of antigen-specific CD4+ T cells within this compartment.
Ly6Chi monocytes recruited to sites of inflammation play a key role in the control of infections due to bacteria (e.g. Listeria monocytogenes), parasites (e.g. Toxoplasma gondii) and fungi (e.g. Cryptococcus neoformans, Aspergillus fumigatus and H. capsulatum) (Serbina et al., 2008; Serbina et al., 2003). Upon arrival in tissue, Ly6Chi inflammatory monocytes have the ability to differentiate into Mϕ and inflammatory dendritic cells (DC), including a subset termed TipDC (TNF-α- and iNOS-producing dendritic cells) (Serbina et al., 2003). The potent immune modulatory effects of TNF-α, and powerful killing products generated with iNOS make these cells forceful effectors against invading pathogens. Thus, a paucity of Ly6Chi monocytes at sites of inflammation during host: pathogen interactions could undermine immunity in ways beyond the failure to prime antigen-specific T cells.
Ly6Chi monocytes exit the bone marrow in response to soluble C-C chemokine signals received through their G-protein coupled receptor CCR2 (Serbina et al., 2008). The chief signals in mice are CCL2, CCL7 and CCL12. We previously reported that CCL7 was elevated in the serum of mice that received the vaccine strain of Blastomyces at the respiratory mucosa (Wuthrich et al., 2012). However, naïve CCR2+Ly6Chi monocytes failed to migrate in response to these sera in vitro, implying that the C-C chemokine was not functional. Likewise, Ly6Chi cells from vaccinated mice migrated poorly in response to recombinant CCL7, implying desensitization of CCR2, which was supported by reduced calcium flux in response to ligand-induced triggering of CCR2 in the cells. We found evidence that vaccine induction of lung matrix metalloproteinase-2 (MMP2) and its action on CCL7 accounted for blunted recruitment of Ly6Chi cells and failed priming of CD4 T cells.
MMP2 is capable of cleaving five residues from the N-terminus of CCL7, which inactivates the chemokine, converts it to an antagonist on CCR2, and desensitizes the receptor (Ali et al., 2005). We found that chemical inhibition of MMP2, vaccination of MMP2−/− mice, and delivery of CCL7 to mice helped vaccine priming of T cells in the lungs (Wuthrich et al., 2012). However, we could not induce levels of CD4+ T cell priming achievable by yeasts that strongly recruit Ly6Chi cells into the lungs, e.g. H. capsulatum. Our findings implied that Blastomyces products, in addition to host MMP2, may also blunt recruitment of Ly6Chi cells to the lungs. If so, such factors could impact the host: fungal pathogen interaction and contribute to immune dysregulation and progressive infection.
Microbes produce extracellular proteases that mediate virulence (Ingmer and Brondsted, 2009). Here, we investigated whether a Blastomyces serine protease blunts influx or function of leukocytes at sites of inflammation, and impacts pathogenesis of disease. We asked 3 questions: i) is yeast viability needed to modulate leukocyte recruitment and, if so, what products mediate the action; ii) what role is played by the serine protease di-peptidlyl peptidase IVA (DppIVA) in modulating leukocyte influx into the lungs, inflammation and virulence of B. dermatitidis; and iii) how might DppIVA exert its action in blunting the recruitment and function of leukocytes.
We report that B. dermatitidis releases extracellular DppIVA, which curtails the influx of Ly6Chi cells into the lungs and impairs the downstream effector functions of these and other leukocytes required for innate defense. We show that DppIVA targets and cleaves C-C chemokines and GM-CSF, which has deleterious consequences for the control of infection. Our work establishes a prominent and unappreciated role for microbial DppIVA in pathogen virulence, involving modes of action that mimic the mammalian enzyme CD26, an ectopeptidase known to modulate critical aspects of hematopoiesis. Aminopeptidases are widely conserved among pathogenic microbes, including bacteria, parasites and fungi, and may represent a strategy by which pathogens undermine mammalian innate host defenses.
RESULTS
A yeast factor blunts leukocyte recruitment
We explored the role of fungal factors in failed recruitment during primary infection. Live, wild type yeast blunted the recruitment of Ly6Chi CCR2+ monocytes, but heat killing the yeast led to an increase in Ly6Chi CCR2+ monocyte recruitment (Fig. 1a), suggesting that a heat-labile factor from the fungus contributes to blunted recruitment of Ly6Chi CCR2+ monocytes. We assessed whether CCR2+ cells are required to control infection and found that the fungal burden is one to two logs higher in Ccr2−/− mice vs. wild-type mice by seven days after infection (Fig. 1b). Thus, blunting the recruitment of CCR2+ cells enhances the virulence of wild-type B. dermatitidis.
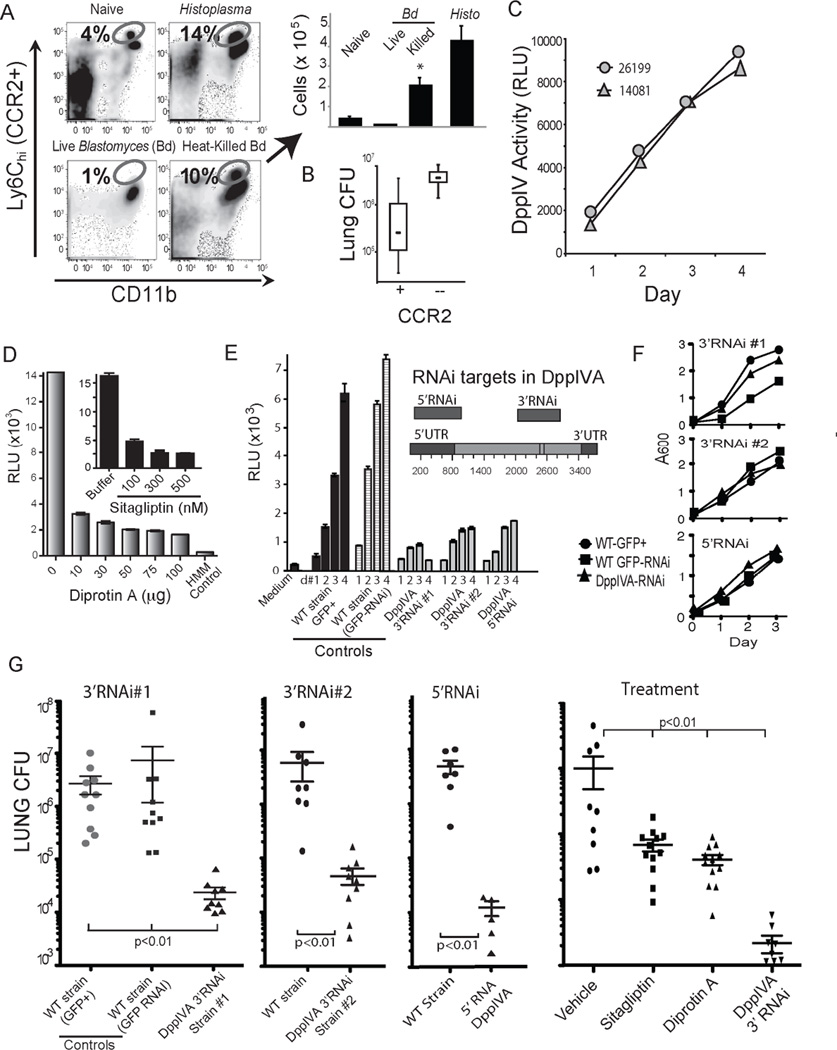
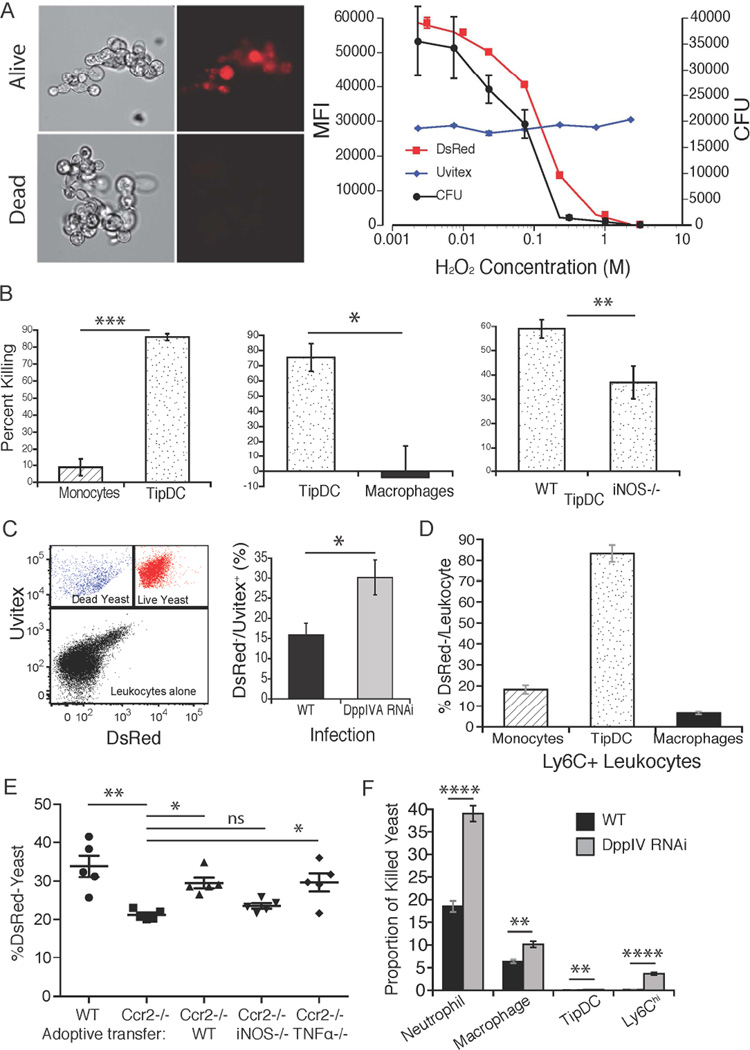
Fig. 1. DppIVA blunts recruitment of Ly6chi CCR2+ monocytes to the lung and promotes B. dermatitidis virulence.
(A) Ly6chi CCR2+ monocytes (circled population) recruited to the lungs of mice 4 days after infection with Histoplasma yeast, or live vs. heat-killed Blastomyces yeast. (B) Lung CFU 7 days post infection in wild-type or Ccr2−/− mice infected with Blastomyces yeast. (C) Glo-assay™ of DppIV activity in culture supernatant of two strains of Blastomyces yeast grown in vitro. RLU, relative fluorescence unit. (D) 7 day culture supernatant of yeast tested for DppIV activity in the presence DppIV inhibitors diprotin A (Ile-Pro-Ile) or Sitagliptin. HMM=medium control. (E) DppIVA 3’ and 5’ regions silenced by RNAi (inset) and DppIV activity (RLU×103) in supernate of isogenic strains: WT (wild-type) GFP+ parental, WT-GFP (only)-RNAi, and DppIVA-RNAi yeast. (F) Growth rate of isogenic DppIVA-silenced and control strains. (G) Lung CFU of mice infected with WT GFP+ parental and RNAi silenced yeast 2 weeks post infection. Right panel shows lung CFU 7 days post-infection in mice that received WT yeast plus indicated treatment. 6–10 mice/group; representative of 3 experiments. (See also Figure S6.)
Mammalian DppIV can cleave and inactivate chemokines and other cytokines responsible for the recruitment and activation of multiple leukocyte populations due to the presence of target proline or alanine residues at the penultimate position of these signals (Boonacker and Van Noorden, 2003). We identified two DppIV homologs in the B. dermatitidis genome – DppIVA and DppIVB; the former harbors a secretion signal and is likely secreted, while the later has a predicted transmembrane domain and is presumed to be intracellular. We tested yeast supernatant for DppIV enzymatic activity and detected activity that increased during growth in vitro (Fig. 1c). Selective DppIV inhibitors DiprotinA and Sitagliptin blocked the generation of signal, indicating that the supernatant activity is due to DppIV (Fig. 1d). Mice also express DppIV (CD26), but B. dermatitidis yeast suppress phagocyte CD26 expression, while mice exposed to wild-type yeast mount an antibody response to fungal DppIV in vivo (Fig. S1a+b), suggesting that the fungal product also is expressed during infection.
DppIVA and fungal virulence
We silenced expression of DppIVA and DppIVB in B. dermatitidis yeast. We generated silenced strains using a GFP-RNAi sentinel system for gene silencing (Krajaejun et al., 2007). Briefly, a GFP+ strain is transformed with a plasmid harboring inverted hairpins for RNAi silencing of both the gene target and the sentinel GFP. The resulting DppIVA RNAi strain showed reduced expression of GFP and the gene of interest; the DppIVB RNAi strain was unstable and GFP silencing could not be maintained. We silenced DppIVA by targeting the 3’ region and 5’ region and confirmed that DppIVA-silenced strains had reduced DppIV enzymatic activity (Fig. 1e). The growth rate in vitro of DppIVA RNAi strains was comparable to isogenic wild-type GFP+ strain and GFP− RNAi strains (Fig. 1f). In a murine model of infection, lung CFU was reduced by 2 to 3 logs with DppIVA RNAi yeast vs. wild-type controls (Fig. 1g). Similar results were observed for multiple independent DppIVA tranformants silenced at the 3’region or 5’region. No perturbations in expression of DppIVB transcript were detected by RT-PCR in DppIVA RNAi strains (data not shown). Thus, silencing DppIVA at multiple regions within the gene, and in multiple independent transformants, attenuated pathogenicity. Likewise, pharmacologic inhibition of DppIV with Sitagliptin or diprotin A ameliorated infection in mice that received wild-type yeast (Fig. 1g). DppIVA thus appears essential for virulence in B. dermatitidis.
DppIVA and blunted Ly6Chi cell recruitment
We tested a link between B. dermatitidis DppIVA and blunted Ly6Chi cell recruitment during infection. We analyzed CFU and the influx of leukocytes to the lung over a five days post-infection (Fig. 2a–c). Ly6Chi cell recruitment to the lungs was greater in mice infected with DppIVA RNAi yeast than wild-type yeast. These recruitment differences were evident by the second day of infection, (Fig. 2c). The numbers of inflammatory Mϕs and dendritic cells (DC) were also increased in mice infected with the DppIVA RNAi strain even though the lung CFU was significantly lower in these mice (Fig. 2c).
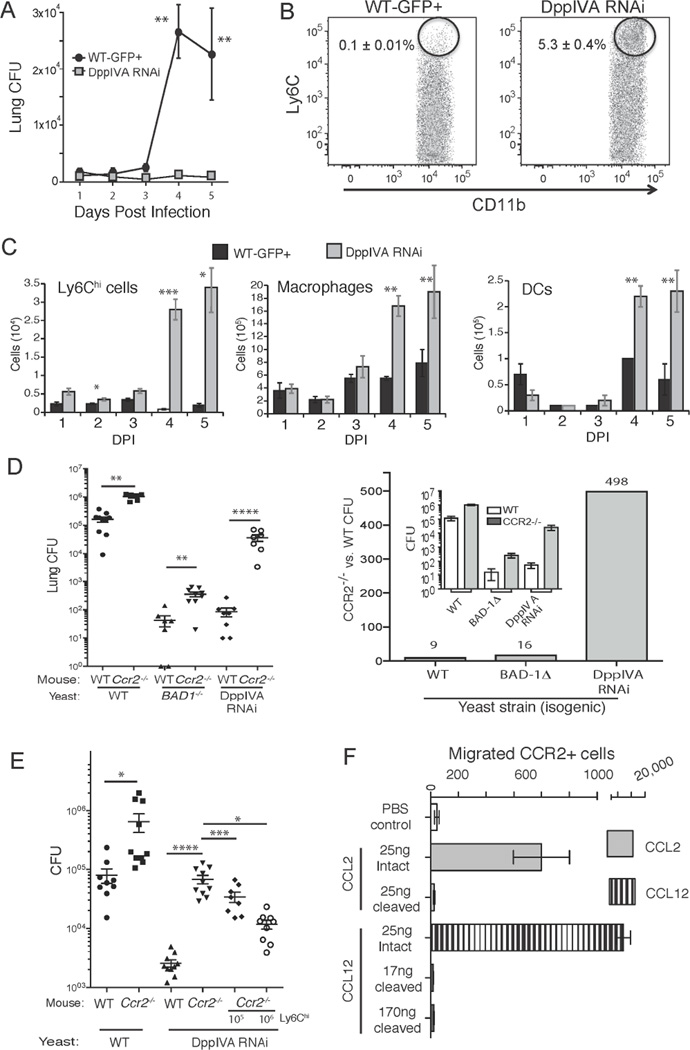
Fig. 2. The inflammatory response to yeast that express DppIVA and the role of CCR2+ monocytes.
(A) Lung CFU of mice infected with isogenic DppIVA-sufficient and –deficient yeast over a 5-days. (B) Percentage of Ly6Chi CCR2+ cells in the lungs 3 days post-infection (events shown are those from the CD11b+ Ly6G− gate) and (C) total number of Ly6Chi cells in lungs. Also shown are the numbers of lung exudative Mϕs and dendritic cells (DC). Data are mean±SEM of 5 mice/group and representative of 3 experiments. (D) Wild-type and Ccr2−/− mice were infected with isogenic yeast and analyzed for lung CFU 2 weeks post infection (left). The mean CFU data (inset) is expressed as fold-change between Ccr2−/− and wild-type mice (right). (E) Ly6Chi cells recruited to lungs and regional lymph nodes of wild-type mice (infected with DppIVA RNAi yeast) were transferred into Ccr2−/− mice infected with DppIVA-silenced yeast. Lung CFU is shown 7 days post infection. Data are mean±SEM of 8–10 mice/group. Results are representative of 2 experiments. (F) Transwell migration of CCR2+ cells in response to intact or fungal DppIVA-cleaved C-C chemokines. Migration was quantified after 16–24 hr of incubation. Results are representative of two experiments. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (See also Figure S1)
Ly6Chi monocytes are recruited to sites of infection via CCR2 (Serbina et al., 2008). To test whether DppIVA is functionally linked with Ly6Chi monocyte recruitment, we analyzed the progression of infection of DppIVA-silenced yeast in Ccr2−/− mice. If linked, the attenuation of DppIV-RNAi yeast should be “suppressed” and lung CFU greatly increased in Ccr2−/− mice. The infection with DppIVA RNAi yeast progressed significantly more in Ccr2−/− vs. wild-type mice (Fig. 2d), resulting in a 498-fold increase in CFU. This increase in fungal burden with DppIVA RNAi yeast was much greater than with wild-type yeast or with another (isogenic) attenuated strain of B. dermatitidis (BAD1−/−) (9 to16-fold increases), indicating a link between DppIVA and virulence in Ccr2−/− mice rather than a general impairment of resistance in the mice.
Although Ly6Chi monocyte recruitment to the lungs was blunted during infection in Ccr2−/− mice, adoptive transfer of Ly6Chi cells into Ccr2−/− mice significantly reduced the burden of lung infection with DppIVA RNAi yeast (Fig. 2e). Thus, Ly6Chi cells restrain infection with DppIVA RNAi yeast, which supports a “gene-to-gene” link between DppIVA in the fungus and CCR2 in the host, leading to blunted recruitment of Ly6Chi cells.
Ly6Chi monocytes are recruited to the lungs via CCR2 and its ligands, CCL2, CCL7, and CCL12. We measured these chemokines in the lung, BAL and serum and found that they were similar in mice infected with wild-type vs. DppIVA RNAi yeast (Fig. S1d and data not shown). Thus, differing chemokine levels do not account for blunted cell recruitment, suggesting functional differences in chemokine action between the two groups. We tested this concept by assaying CCR2+ cell migration in response to intact vs. fungal DppIVA cleaved C-C chemokines. Trimming two amino acids from N-terminal of these C-C chemokines, as confirmed by Mass Spectrometry, impaired the ability of CCR2 to induce migration of reporter cells (Fig. 2f) and of primary Ly6Chi cells (Fig. S1c). Lung MMP2 levels were >3-fold higher in mice that received wild-type yeast vs. DppIVA RNAi yeast (data not shown), and pharmacological inhibition of MMP2 further augmented influx of Ly6Chi cells in these mice (Fig. S1c). Thus, our Ly6Chi results can best be explained by cleavage and inactivation of one or more chemokines by DppIVA, together with elevated MMP2 and desensitized CCR2 as described (Wuthrich et al., 2012).
Enzymatic action of fungal DppIVA on GM-CSF and downstream effects
GM-CSF promotes differentiation of Ly6Chi cells into DCs in vivo and in vitro (Hamilton, 2008). We found that inflammatory (Ly6Chi) DCs and TipDCs accumulated in greater numbers in the lungs of mice infected with DppIVA RNAi yeast vs. wild-type yeast (Fig. 3a+b). GM-CSF expression was similarly upregulated during infection with both strains of B. dermatitidis (Fig. 3c). However, GM-CSF harbors a penultimate proline at the N-terminus and is cleaved and inactivated by mammalian DppIV (Broxmeyer et al., 2012). We tested whether B. dermatitidis DppIVA cleaves GM- CSF in vitro. By 6 hr of incubation, DppIVA had fully removed both N-terminal residues from GM-CSF in the presence or absence of serum (Fig. 3d), and digestion was blocked by the DppIV inhibitor diprotin A. Wild-type yeast cultured in serum (without mammalian cells and CD26) also cleaved GM-CSF (Fig. 3e). Since cleaved GM-CSF has diminished action on its receptor (Broxmeyer et al., 2012), the loss of GM-CSF action could have untoward effects in vivo due to impaired differentiation of DC or activation of other leukocytes.
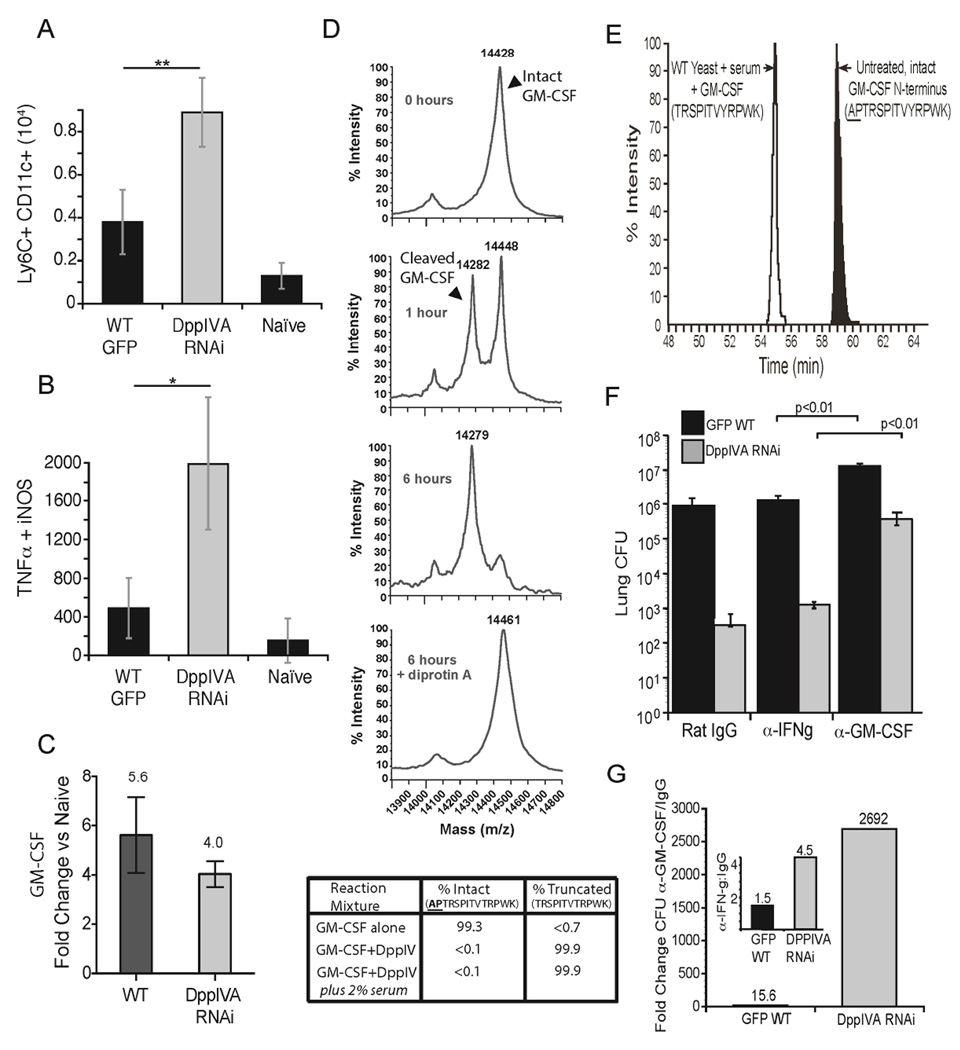
Fig. 3. Action of yeast DppIVA on GM-CSF and impact on evolution of infection.
Number of monocyte-derived DC (CD11c+, Ly6Chi cells) (A) and TipDC (TNF-α, NOS2+, CD11c+, Ly6Chi) (B) in the lungs 4 days post-infection. *p<0.05, **p<0.01. (C) Fold increase in transcript of GM-CSF measured by RT-PCR in lung homogenate 2 days post infection compared to naïve mice. Data in panels A–C are mean±SEM of 3–5 mice/group and are representative of 2 experiments. (D) MS analysis of GM-CSF cleavage by fungal rDppIVA at 0, 1, and 6 hr of incubation alone or with DiprotinA. Table shows extent of removal of N-terminal two amino acids (AP) by rDppIVA in buffer alone or with 2% serum. (E) Wild-type yeast were cultured in vitro in medium with GM-CSF and 2% mouse serum. After 72 hr, GM- CSF was collected by chromatography and analyzed by MS for residues at the N-terminus (open peak) vs. untreated GM-CSF (shaded peak). (F) Mice received rat IgG or neutralizing α-GM-CSF or α-IFN-γ and were infected with yeast strains, and lung CFU enumerated as above in panel E. (G) Fold change in CFU between GM-CSF neutralized vs. rat IgG treated mice is shown for each yeast strain. Inset shows fold-change for rat IgG-treated vs. IFN-γ neutralized mice. Numbers above bars are fold-change. Data in panels E+F are mean ± SEM of 8–10 mice/group, and are representative of 2 experiments. (See also Figure S4).
We tested the link between fungal DppIVA and GM-CSF in vivo during infection. We hypothesized that DppIVA cleavage of GM-CSF in vivo would promote immune evasion by wild-type yeast, and conversely that loss of DppIVA would enhance GM-CSF mediated clearance of a DppIVA RNAi strain. If true, growth of the latter strain would be preferentially enhanced in mice lacking this cytokine. To test this hypothesis, we neutralized GM-CSF during infection (Fig. 3f+g). Lung CFU in mice infected with the DppIVA RNAi strain was sharply elevated compared to infection with wild-type yeast in GM-CSF-neutralized mice. In contrast, neutralization of IFN-γ (not expected to be a substrate of DppIVA) had a negligible effect on infection by both yeast strains (Fig. 3g). In a second approach, we infected GM-CSF receptor knockout (Csf2ra−/−) mice with wild-type or DppIVA RNAi yeast. The absence of GM-CSF receptor signaling in Csf2ra−/− mice significantly increased growth of the DppIVA RNAi strain by 12,098-fold, while growth of the wild-type strain was increased by 1166-fold (Fig. S4a). The increased growth of the wild-type strain in Csf2ra−/− mice suggests incomplete cleavage of GM-CSF by fungal DppIVA in wild-type mice. Thus, DppIVA is functionally linked with GM-CSF in vivo, implying that inactivation of the cytokine during infection with wild-type yeast is essential for pathogen virulence.
Analysis of host: pathogen encounters in vivo with a reporter strain of yeast
We engineered a DsRed reporter strain of B. dermatitidis to track yeast encounters with host leukocytes in vivo and assess their impact on yeast survival i.e. which cells associate with and kill yeast. DsRed signal is strong in viable yeast and absent in dead yeast (Fig. 4a). The DsRed signal correlates with CFU (Fig. 4a+S2a) as reported for A. fumigatus (Jhingran et al., 2012). During co-culture in vitro with DsRed yeast, TipDCs killed better than monocytes or Mϕs, and required iNOS for killing (Fig. 4b+S2b). By using uvitex to stain cell wall chitin and detect all yeast in vivo e.g. live and dead (Fig. 4c), we defined the identity of leukocytes associated with yeast and the ability of different leukocytes to kill them during infection. Dead yeast were more often associated with TipDCs than with Ly6Chi monocytes or Ly6C+ cell derived Mϕs in vivo (Fig. 4d). To test the role of TipDCs as a cellular basis for the loss of immune control of the silenced yeast, we transferred Ly6Chi-derived inflammatory DCs into Ccr2−/− mice infected with DppIVA RNAi yeast. Addition of these DCs (capable of producing iNOS and TNF-α when stimulated with yeast [Fig. S2b and data not shown], enhanced the killing of DsRed yeast in vivo and did so in a manner dependent on iNOS, but not TNF-α (Fig. 4e). Thus, TipDCs are potent effectors in vivo and yeast DppIVA undermines their development during infection. However, proportionately more killing of yeast is mediated by Mϕs and neutrophils in vivo due to their greater absolute numbers in infected lungs, and these populations also are more active against DppIVA RNAi yeast vs. wild-type yeast in vivo (Fig. 4f). We studied each of these leukocyte populations further below.
Fig. 4. A yeast reporter to decipher the host:fungal pathogen encounter in vivo.
(A) B. dermatitidis DsRed reporter shows loss of fluorescence in nonviable yeast (left). Loss of DsRed (mean fluorescence intensity, MFI) over increasing [H2O2] correlates with CFU, while uvitex signal (cell wall chitin) persists (right). (B) DsRed yeast (DppIVA RNAi strain) were cultured in vitro with cells shown at effector: target ratio of 3:1 for 24 hr and CFU recorded to assess percent yeast killed. See similar results for DsRed wild-type in S2b. (C) Fluorescent appearance of Uvitex+, dead (DsRed−) vs. live (DsRed+) yeast in vivo (left), and the proportions of dead yeast 4 days post-infection with wild-type and DppIVA RNAi yeast (right). (D) Percentage of non-viable yeast associated with Ly6Chi monocyte-derived leukocytes 4 days after infection with DppIVA RNAi yeast. (E) Adoptive transfer of in vitro elicited wild-type, iNOS−/− and TNF-α−/− inflammatory DCs (106/recipient) into Ccr2−/− mice infected with DsRed, DppIVA-silenced yeast. (F) Proportion of dead DsRed yeast associated with leukocytes relative to yeast expression of DppIVA. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (See also Figure S2).
DppIVA promotes survival of yeast in GM-CSF activated macrophages
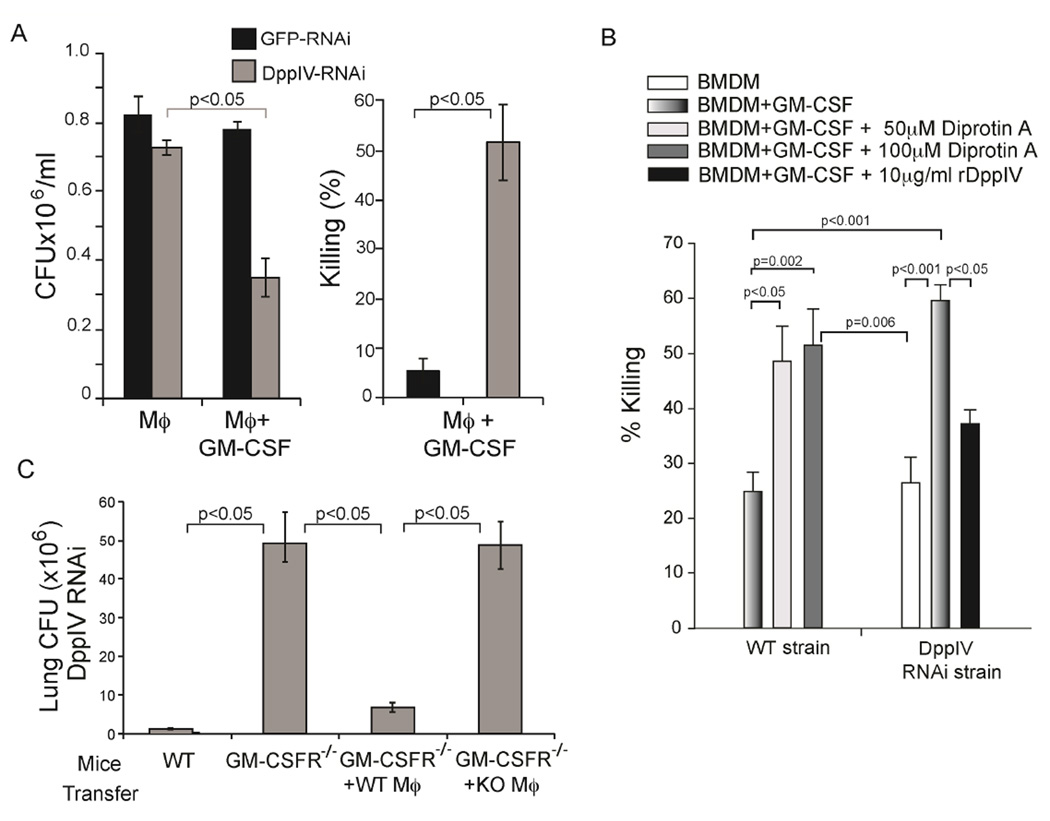
To further assess how DppIVA production by yeast provides a survival advantage to the pathogen, we co-cultured bone marrow-derived Mϕs with wild-type or DppIVA RNAi yeast in the presence of GM-CSF. GM-CSF induced meager growth inhibition of wild-type yeast, but the growth of DppIVA RNAi yeast was inhibited by nearly 60% (Fig. 5a). Moreover, blocking DppIV activity with diprotin A boosted growth inhibition of wild-type yeast to levels near those observed for DppIVA RNAi yeast (Fig. 5b). To test whether addition of DppIVA would rescue growth of DppIVA deficient RNAi yeast, we supplemented culture media with recombinant (r) Blastomyces DppIVA. Addition of rDppIVA reversed the growth inhibitory capacity of GM-CSF activated Mϕs and improved the survival of DppIVA RNAi yeast (Fig. 5b). GM-CSF activated Mϕs from human donors yielded similar results (Fig. S3a). Thus, DppIVA produced by yeast counteracts the anti-fungal effect of GM-CSF activated Mϕs.
Fig. 5. GM-CSF enhances killing of yeast by macrophages that is reversed by fungal DppIVA.
(A) Yeast CFU (left) and percent killing (right) of yeast cultured for 24 hr with GM-CSF activated bone marrow derived Mϕs (BMDM). (B) Growth inhibition of yeast cultured with GM-CSF activated BMDM treated with DppIVA or diprotin A. (C) Lung CFU in Csf2ra−/− mice 7 days after infection with DppIVA RNAi yeast and adoptive transfer of wild-type vs. Csf2ra−/− peritoneal Mϕs. (See also Figure S3).
Our in vitro studies revealed that yeast DppIVA impedes the anti-fungal action of Mϕs in the presence of GM-CSF. To validate that GM-CSF regulates the response of Mϕs to infection in vivo, we adoptively transferred peritoneal Mϕs from wild-type or Csf2ra−/− mice into Csf2ra−/− mice just prior to infection with the DppIVA RNAi strain. The transfer of wild-type Mϕs into Csf2ra−/− mice sharply enhanced antifungal immunity, and decreased growth of the DppIVA RNAi strain in Csf2ra−/− mice (Fig. 5c). In contrast, transfer of Csf2ra−/− Mϕs failed to curtail growth of this fungal strain. Adoptive transfer of wild-type Mϕs to Csf2ra−/− mice helped control infection with wild-type yeast as well, but not to the same extent as with DppIVA RNAi yeast (Fig. S4b). Csf2ra−/− mice possess increased amounts of pulmonary GM-CSF due to lack of signaling via the GM-CSF receptor (Carey and Trapnell, 2010). Introduction of wild-type Mϕs in these mice presumably restored GM-CSF signaling in the transferred cells. Thus, GM-CSF activation of Mϕs helps control a dimorphic fungus in vivo, and GM-CSF cleavage and inactivation by DppIVA from wild-type yeast appears to promote fungal escape from Mϕ-mediated killing and progression of disease.
Impact of DppIVA inactivation of GM-CSF on neutrophils
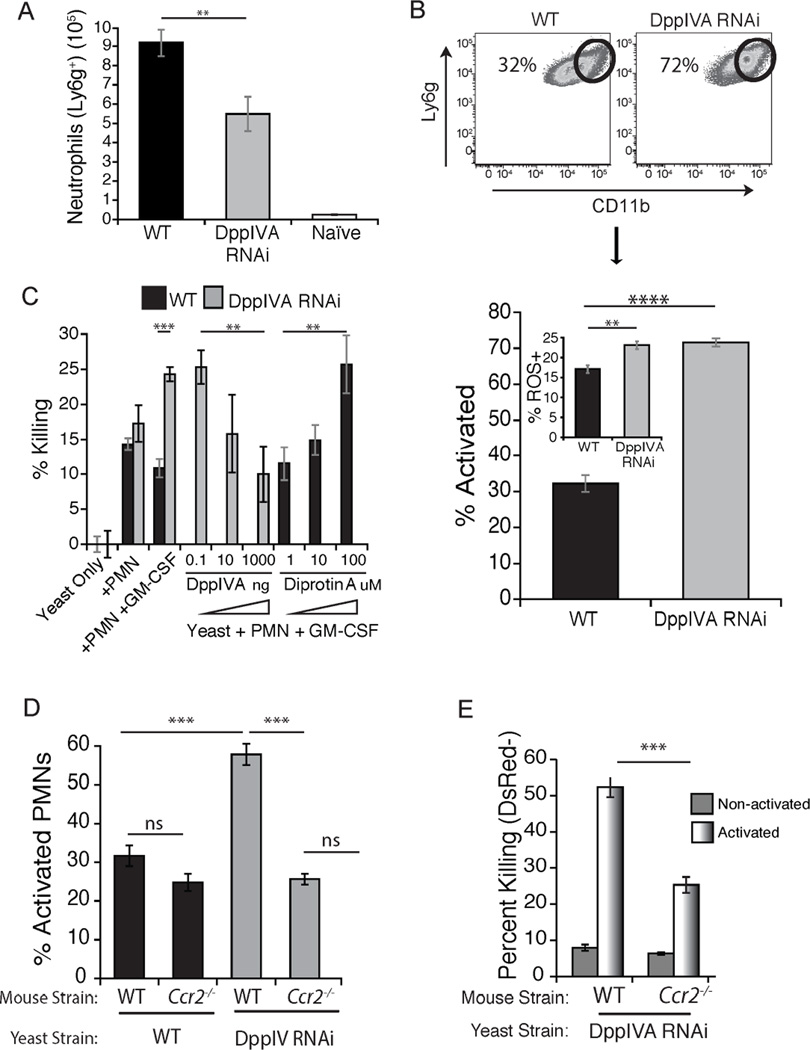
GM-CSF is a polyfunctional cytokine; it activates neutrophils and augments their anti-fungal action (Hamilton, 2008). We hypothesized that fungal DppIVA cleavage of GM-CSF would curtail neutrophil activation and killing of yeast. We found that the number of neutrophils recruited to the lungs during infection was higher in response to wild-type vs. DppIVA RNAi yeast (Fig. 6a), and correlated with higher lung CFU by day 4 of infection, although the ratio of neutrophils to yeast was lower in wild-type infection (Fig. S5b). Likewise, a much smaller proportion of the cells displayed an activated phenotype in mice infected with wild-type vs. DppIVA RNAi yeast, as measured by surface expression of CD11b or CD11a and Ly6G (Figs. 6b+S5a). Similarly, the two groups of mice showed corresponding differences in neutrophil ROS production, with less ROS production in the neutrophils from mice infected with wild-type yeast.
Fig. 6. Neutrophil action against yeast is blunted by fungal DppIV and requires both GM-CSF and monocyte recruitment.
(A) Number of neutrophils recruited to the lung at day 4 post-infection. Data are mean±SEM of 3–5 mice/group and representative of 2 experiments. (B) Percentage of neutrophils that are activated (Ly6G+ and CD11bhi) in infected mice. Dot plot of lung neutrophils is from a representative mouse (top) and histogram (bottom) is from 10 mice/group at 4 days post infection. Inset shows the percentage of ROS+ lung neutrophils without ex vivo stimulation in these mice. (C) Percentage of yeast killed after culture for 6 hr with neutrophils activated with GM-CSF. Some wells contained fungal rDppIVA or DppIV inhibitor diprotin A. (D) Percentage of lung neutrophils activated (CD11bhi) at four days after infection, illustrating how impaired CCR2+ monocyte recruitment blunts activation of neutrophils and killing (E) of yeast. Data are the mean ± SEM of 3–5 mice/group and representative of 2 experiments. **p<0.01, ***p<0.001, ****p<0.0001. (See also Figures S5 and S6).
We tested the roles of GM-CSF and fungal DppIVA in neutrophil mediated killing of yeast in vitro. Addition of GM-CSF to neutrophils in vitro enhanced their killing of DppIVA silenced yeast, but did not affect the killing of wild-type yeast (Fig. 6c); killing required GM-CSF receptor on the neutrophils (Fig. S4c). Addition of fungal rDppIVA prevented the killing of DppIVA silenced yeast by GM-CSF treated neutrophils, lowering the level of killing (10%) to that observed for non-treated neutrophils against wild-type yeast (Fig. 6c). Conversely, addition of the DppIV inhibitor, DiprotinA, increased the ability of GM-CSF treated neutrophils to kill wild-type yeast (Fig. 6c). Diprotin A enhanced killing of wild-type yeast in a concentration dependent manner, yielding levels of killing (25–30%) similar to that observed by GM-CSF treated neutrophils against the DppIVA silenced strain. Neutrophils from human donors mirrored results with murine cells (Fig. S3b). Of note, rDppIVA did not directly affect or reduce neutrophil killing of the silenced strain; it did so only in the presence of GM-CSF (Fig. S5b). The addition of DiprotinA reversed the ability of rDppIVA to suppress killing of silenced yeast by GM-CSF treated neutrophils (Fig. S5b). Neither soluble DppIVA nor DiprotinA affected in vitro growth of yeast in the absence of phagocytes (Fig. S5c). Thus, fungal DppIVA blunts priming of neutrophils by inactivating GM-CSF, promoting survival of yeast and disease progression.
In murine Aspergillosis, CCR2+ monocytes augment neutrophil killing of conidia (Espinosa et al., 2014). We found that the blunted activation state of neutrophils in Ccr2−/− mice lacking CCR2+ monocytes mirrored that of wild-type mice infected with wild-type yeast, in which monocyte recruitment is blunted (Fig. 6d). Likewise, although the killing of yeast in vivo in Ccr2−/− mice is higher in activated neutrophils than in non-activated neutrophils, killing by the former is nevertheless sharply impaired in the absence of recruited CCR2+ monocytes (Fig. 6e). Adoptive transfer of Ly6Chi monocytes into Ccr2−/− mice significantly enhanced the killing of DppIVA RNAi yeast in vivo by neutrophils and neutrophil-DC hybrids, as well as by Mϕs (Fig. S6). Thus, Ly6Chi monocytes not only can kill the fungal pathogen directly (Fig. 4b+c), and can differentiate into Mϕs and DCs that are even more potent killers, but Ly6Chi monocytes also appear to provide signals to neutrophils and other phagocytes that augment their ability to kill the fungal pathogen.
DISCUSSION
The dimorphic fungi exert profound immune regulatory disturbances during the course of progressive, systemic infection, but little is known about the molecular basis of these defects. We report that fungal DppIVA subverts elements of host innate immunity in a murine model of pulmonary infection with B. dermatitidis, and accounts for progressive, lethal infection. Conversely, elimination of expression of DppIVA in the pathogen attenuates its virulence and is associated with restoration of early leukocyte recruitment, function and control of disease. Thus, DppIVA is strongly implicated in these events.
Gene silencing may have off target effects, limiting the strength of our conclusions. To guard against confounding effects, we took several measures. First, we silenced multiple regions of DppIVA, including at the 3’ and 5’ ends. Second, we avoided targeting regions of homology with DppIVB. Third, we ensured that DppIVB transcript was unaffected in DppIVA-silenced strains and that enzyme activity was specifically reduced in DppIVA silenced strains. Fourth, to control for random integration of silencing constructs, we studied multiple, independent transformants of DppIVA silenced strains. Fifth, we controlled for the effects of RNAi-silencing machinery by including strains in which only GFP was silenced (i.e. transformants where the transforming DNA had only GFP in the RNAi cassette), and showed that these strains have wild-type levels of DppIV activity and virulence. Nevertheless, we were unable to obtain a clean, targeted deletion of the DppIVA gene in B. dermatitidis, offering unambiguous proof. Still, the immune disturbances identified in our study are highly reminiscent of the actions ascribed to mammalian DppIV.
DppIV is a multifunctional protein conserved in mammals and microbes. DppIV is a member of the prolyl oligopeptidase family known for its function as a serine protease, selectively cleaving the N-terminal penultimate proline or alanine from proteins. The best studied DppIV is human CD26, which removes dipeptides from chemokines, binds extracellular matrix constituents such as collagen and fibronectin, and interacts with other proteins such as adenine deaminase on T cells and a Na+/H+ antiporter pump on kidney cells (Boonacker and Van Noorden, 2003). Human DppIV cleaves CXCL10 (IP-10) and CXCL12, inactivating these attractants of T cells and hematopoietic stem cells, respectively (Ou et al., 2013). Over 40 cytokines and chemokines display a penultimate proline or alanine and are potentially cleavable by DppIV (Ou et al., 2013). Cleavage of such targets by microbial DppIV could foster immune evasion, but to our knowledge, there are no firm examples of this microbial evasion strategy.
We found that recruitment of Ly6Chi CCR2+ monocytes into the lungs was blunted during infection with wild-type yeast, but not DppIVA silenced yeast. The C-C chemokine signals that attract these cells to sites of inflammation include CCL2, CCL7 and CCL12. The lung and serum levels of transcript and protein for these products did not differ in mice infected with these strains, implying a difference in their functional activity in the two settings. Each of these targets displays a penultimate proline target of DppIV cleavage. Removal of several N-terminal residues impairs the activity of these chemokines on CCR2, converts them into antagonists, and desensitizes CCR2 itself.
Pathogenic yeasts may prevent monocyte recruitment through mechanisms other than DppIVA cleavage of CCR2 ligands. DppIVA may indirectly upregulate MMP-2, which can block Ly6Chi monocyte recruitment by cleaving CCL7 (Wuthrich et al., 2012). DppIV can act on M6P/IGF-IIR to induce ROS production from epithelial cells (Ishibashi et al., 2013) and ROS can induce MMP-2 and MMP-9 production (Rajagopalan et al., 1996). We observed a 3 to 4-fold increase in lung MMP-2 transcript and product in mice infected with wild-type yeast vs. DppIVA RNAi yeast. Elevation in lung MMP2 likely contributes to impaired CCL7 function, as we observed with vaccine strain delivery at the respiratory mucosa (Wuthrich et al., 2012).
Fungal DppIV might also impair the migration of leukocytes during wild-type infection due to an impaired chemokine gradient. Mammalian DppIV binds fibronectin and collagen and promotes MMP-mediated digestion of extracellular matrix (ECM) (Boonacker and Van Noorden, 2003). Chemokines presented on ECM establish a gradient that facilitates leukocyte migration. DppIV enhanced destruction of ECM may therefore alter this gradient and impair the normal physiology of leukocyte migration.
One of the most profound effects of fungal DppIVA is its action on mammalian GM-CSF. This cytokine has many functions during the generation of an immune response to pathogens, involving the differentiation and activation of monocytes, Mϕs, DCs, and neutrophils (Hamilton, 2008). GM-CSF cleavage by mammalian DppIV inactivates the cytokine and the cleaved form acts as a high-affinity, competitive inhibitor of intact GM-CSF on its receptor (Broxmeyer et al., 2012). We found that fungal DppIVA likewise cleaves GM-CSF. During in vitro experiments, the expression of DppIVA by yeast protected the pathogen from leukocyte killing by undermining GM-CSF activation of both murine and human Mϕs and neutrophils. Likewise, we detected a relative paucity of TipDCs during infection with wild-type yeast and found that these immune cells, when provided GM-CSF during their development, effectively kill the fungus. GM-CSF’s role in defense against pathogens is complex and its inactivation may have additional effects beyond what we describe here including DC and Mϕ differentiation, leukocyte adhesion, and cytokine production (Hamilton, 2008).
We engineered DsRed reporter yeast to monitor host: pathogen encounters in vivo in the lung during infection. Although TipDCs were potent at killing yeast in vitro and in vivo, these immune cells accounted for only a small fraction of dead yeast in the lung compared to Mϕs and especially neutrophils. Neutrophil are a signature feature of the granulomatous inflammatory response to Blastomyces, accounting for the characteristic pyogranuloma. Although these cells were most often associated with dead yeast, presumably due to their effector function, there was a striking difference in neutrophil killing of the two fungal strains in vivo, with many more viable wild-type yeast than DppIV silenced yeast. We linked this difference to impaired activation and ROS production by neutrophils in vivo. Since treatment of neutrophils with GM-CSF primes and activates them and enhances their ROS production, it is likely that inactivation of GM-CSF by DppIVA contributes to the defect. However, the blunted influx of CCR2+ monocytes that we observed – in the setting of DppIVA production or in Ccr2−/− mice – also impaired the activation of neutrophils and their in vivo killing of the fungal pathogen. During murine pulmonary Aspergillosis, CCR2+ monocytes and monocyte-derived TipDCs promote neutrophil killing of Aspergilllus conidia through a mechanism of cross-talk that augments fungicidal action of neutrophils (Espinosa et al., 2014). Although neutrophils pour into the lesions of Blastomyces pneumonia, the cells appear to be deprived of the activation signals – GM-CSF and possibly other monocyte-derived signals – needed to eliminate the fungus. Thus, yeast DppIVA acts in at least two ways to foster fungal survival in vivo in the face of neutrophils: by inactivating GM-CSF and impairing recruitment of CCR2+ monocytes and their communication with neutrophils. At least 94 chemokines, cytokines, and immune-modulatory factors are potential targets for DppIV (Ou et al., 2013). Thus, fungal DppIVA cleavage and inactivation of GM-CSF may only be part of its function.
Our study focused on pathogen evasion of innate immunity. However, DppIVA may also undermine adaptive immunity. GM-CSF regulates differentiation of DCs, which bridge innate and adaptive immunity. We previously reported impaired adaptive immunity in response to fungal vaccination in the lung, and attributed this defect to exuberant MMP2 and blunted recruitment of inflammatory monocytes (Wuthrich et al., 2012). The combined effect of DppIVA and MMP2 on blunting the number and function of monocyte-derived DC could together undermine priming of antigen-specific T cells. DppIV could also modulate adaptive immunity by cleaving and inactivating T cell-activating cytokines such as TGF-β, IL-17 and IL-23 or products such as IL-2 (Ou et al., 2013). The many potential DppIV targets and effects their cleavage may have on the immune response to infection indicates multiple ways in which the fungus may dysregulate immunity.
The discovery of fungal DppIVA as a virulence factor makes it a potential therapeutic drug target. The DppIV inhibitor Sitagliptin inhibited fungal DppIV activity in vitro and drug treatment ameliorated disease in vivo. Sitagliptin is one of several DppIV inhibitors and was initially approved for human use to treat type II diabetes in 2006 (Yazbeck et al., 2009). DppIV inhibitors have been investigated for their ability to treat multiple diseases including arthritis, multiple sclerosis and inflammatory bowel disease (Yazbeck et al., 2009). Perhaps inhibitors of DppIV could be repurposed as therapeutic adjuncts for fungal or other infections in which DppIV is found to promote microbial pathogenicity.
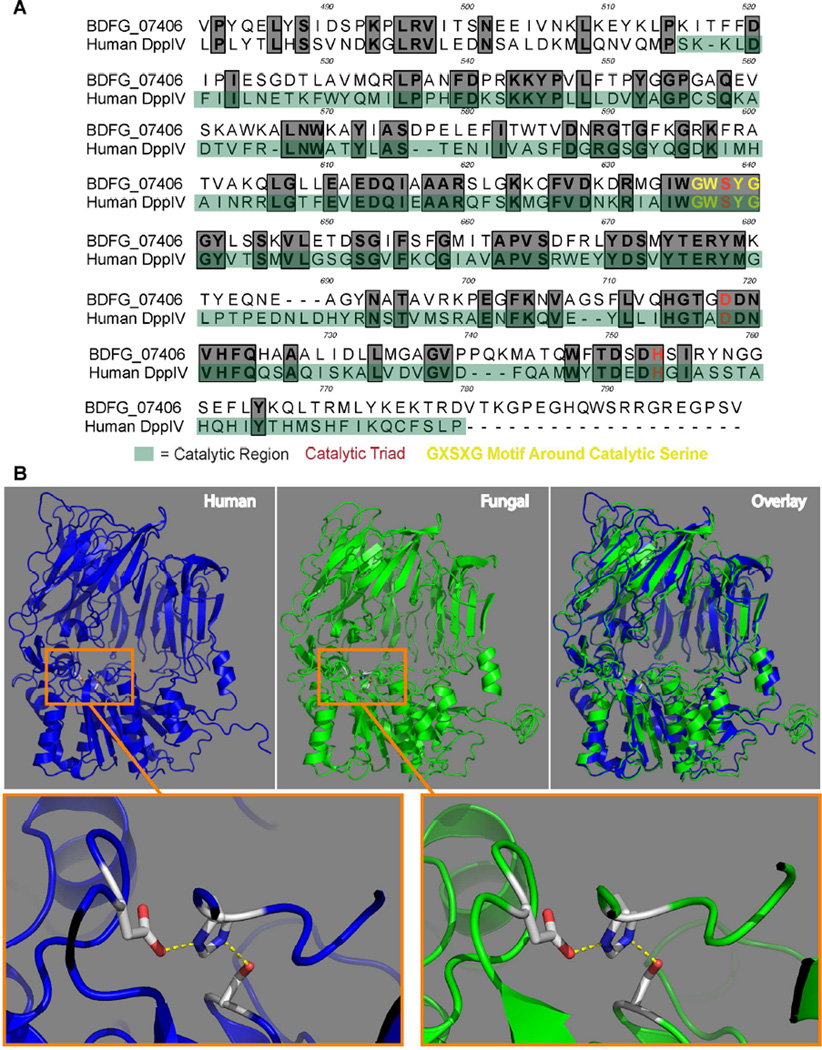
DppIV is conserved in mammals and microbes. We compared mammalian and fungal DppIV and found that the amino acid sequences and 3D structures are similar to one another, especially around the active site for catalytic cleavage (Figs. 7 and S7). Both of enzymes likewise cleave at least one substrate similarly, GM-CSF. Many of separate protease families may have evolved by convergence, but for individual subgroups, including some di-peptidyl peptidases, the genes across phyla seem to have evolved from a common ancestor (Qi et al., 2003). To our knowledge, DppIV has not been described as a fungal virulence factor. DppIV is expressed in Aspergillus, but has not been studied for a role in pathogenesis. CCL2 and CCR2+ cells promote resistance to this pathogen (Espinosa et al., 2014; Morrison et al., 2003) reminiscent of their roles with Blastomyces. In H. capsulatum, gene deletion of DppIVA did not cause a virulence defect (Cooper and Woods, 2009). However, DppIVB is responsible for most of the extracellular DppIV activity and has not been tested for a role in pathogenicity in vivo (Cooper et al., 2009). DppIV has been implicated in the pathogenesis of other infectious diseases such as periodontal disease from Porphyromonas gingivalis, Streptococcal toxic shock, and liver disease from Hepatitis C (Casrouge et al., 2011; Ge et al., 2009; Kumagai et al., 2000). Similar peptidases have been studied for their role in Giardia encystation, Trypanosome pathogenesis, and malarial release from RBCs (Antoine-Moussiaux et al., 2009; Arastu-Kapur et al., 2008; Touz et al., 2002). The immune disturbances and progressive disease attributed here to apparent fungal mimicry of mammalian DppIVA, in conjunction with the conservation of this ectopeptidase throughout the microbial kingdom, suggest an unappreciated mode of molecular piracy among microbial pathogens.
Fig. 7. Comparison of B. dermatitidis and human DppIV.
(A) Primary amino acid sequence alignment of the catalytic region of B. dermatitidis DppIVA (Gene ID: BDFG_07406, Uniprot ID: T5BJB9) and human DppIV (Uniprot ID: P27487), which are 44% identical and 59% similar as determined by NCBI blastp (protein-protein BLAST). Identical amino acids are bolded and boxed in gray. (B) (Top Left) 3D Model of the previously crystalized human DppIV protein with the catalytic triad (aspartic acid, histidine, serine) boxed. (Top Center) Predicted 3D structure of B. dermatitidis DppIVA generated in I-TASSER with the catalytic triad (aspartic acid, histidine, serine) boxed. (Top Right) Overlay of 3D structures of human and B. dermatitidis DppIV. (Bottom Left) Zoom-in of the human DppIV catalytic triad (Bottom Right) and B. dermatitidis DppIV catalytic triad. All 3D models were made using PyMOL (The PyMOL Molecular Graphics System). (See also Figure S7)
METHODS
Mice
C57BL/6 wild type mice were obtained from NCI or Jackson Laboratory. Ccr2−/− mice were obtained from Jackson and bred in house. Csf2ra−/− mice were provided by Bruce Trapnell, University of Cincinnati.
Fungi
We used B. dermatitidis ATCC strain 26199 or 14081 and H. capsulatum ATCC strain G217B. See Supplement for growth of strains.
GFP-sentinel silenced strains
DppIV-silenced (RNAi) and control strains were engineered with the B. dermatitidis GFP-sentinel system (Krajaejun et al., 2007). Two non-overlapping sequences of DppIV at the 5’ region and 3’region (BDFG_07406) were targeted for silencing. See Supplement for detail. Vectors for silencing DppIV or GFP were transformed into Agrobacterium tumefaciens. A. tumefaciens-mediated transformation (AMT) (Sullivan et al., 2002) was used to transform the 26199-GFP strain with non-silencing or silencing plasmids to respectively generate a non-silencing control strain and DppIV-silenced strains. Silenced transformants were identified by loss of GFP fluorescence and confirmed by DppIV-Glo Protease Assay (Promega).
DsRed strains
Agrobacterium binary vector construction is described in Supplement. Agrobacterium- transformation of B. dermatitidis strain 26199 with pCAMDsRed#2 and selection for hygromycin resistance were done as described (Sullivan et al., 2002). Transformants were screened for DsRed expression with a BioRad Versadoc 5000 (Green LED, 605 nm filter). Red fluorescent colonies were streaked on selection medium and isolated colonies screened by Versadoc and analyzed by fluorescent microscopy and flow cytometry to confirm uniformly fluorescent cells.
Infection
Mice were were anesthetized with isoflurane, suspended by their front incisors from a wire and infected by intubation using a BioLite instrument (Braintree Scientific). Yeast were given intratracheally (i.t.) by cannula. Some mice were infected i.t. by surgery. Infections were done with 2 to 3×104 yeast in 20 µL of PBS per mouse unless noted. Higher inocula were used to assay yeast association with small leukocyte populations or populations with low association with yeast in the lung.
Macrophages (Mϕs)
Marrow was collected from femurs and tibias by rinsing and disruption via a 26-gauge needle and filtered via a 40 µM filter. Mϕs were differentiated as in Supplemental methods and co-cultured with yeast at a multiplicity of 1:1. 24 hr post infection, Mϕs were lysed in hypotonic buffer (20 mM Tris HCl, 10 mM NaCl, 3 mM MgCl2, pH 7.5) and yeasts plated on HMM agar.
Neutrophils
Neutrophils were purified from bone marrow of 6–8 week old wild-type mice. Wells contained ratios of neutrophils: yeast of 50 to 100: 1. Some wells received 10 ng/mL GM-CSF. Co-cultures were incubated for 6 hr at 37°C and 5% CO2. Each condition had 6–8 replicates. After cell lysis, yeast were plated on HMM. Percent killing was defined as (1-[# of yeast in a condition/# of yeast without neutrophils]) × 100. Neutrophils were depleted with 250µg α-Ly6g (clone 1A8) (BioXCell) given i.v. every other day.
Ly6Chi cells
Ly6Chi cells were purified from bone marrow using a 2-step approach on the AutoMACS system (Miltenyi) described in Supplemental methods. Ly6C-derived cells were confirmed by flow cytometry. TipDCs generated from Ly6Chi derived inflammatory DCs produced nitric oxide and TNF-α in response to exposure to yeast (Fig. S2b and data not shown) as measured by Griess reaction and/or respective transcript, consistent with TipDCs (Serbina et. al., 2003). To test yeast killing in vitro by Ly6C-derived cells, cells were cultured with yeast for 24 hr at 37° C (without GM-CSF) at a ratio of 3:1. For adoptive transfer into Ccr2−/− mice, Ly6C+-derived cells were transferred as noted in Supplemental methods.
Csfra2−/− cells and experiments
Peritoneal Mϕs were obtained by lavage of mice with Hank’s balanced salt solution (HBSS) (Corning). Details are in the Supplement. With DppIVA RNAi yeast, 1.5 × 106 peritoneal Mϕs from wild-type or Csf2ra−/− mice were transferred i.t. into 8–12 week old Csf2ra−/− mice just prior to i.t. infection with 3×104 yeast. With wild-type yeast, 0.5 × 106 peritoneal Mϕs from wild-type or Csf2ra−/− mice were transferred i.t. into 8–12 week old Csf2ra−/− mice just prior to i.t. infection with 3×104 yeast.
GM-CSF neutralization
Mice were given 250 µg of antibody i.v. every other day. We used mAb XMG1.2 (rat IgG1 α-IFN-γ) provided by R. Seder (NIH) and MP1-22E9 (rat IgG2a anti-GM-CSF) with permission of J. Abrams (DNAX). Ascites was made in BALB/c Nu/Nu males. IgG was (NH4)2SO4 precipitated and quantified by OD280. Controls received rat IgG (Sigma).
Recombinant DppIVA
Expression of B. dermatitidis rDppIVA was performed using EasySelectPichia Expression Kit (Invitrogen). See Supplement for detail.
DppIV GLO Assay
DppIV Glo-Protease assays were performed in a 96-well white-walled, clear bottom plate as per the manufacturer’s protocol (Promega). Details are in Supplement methods.
Statistical Analysis
p values were calculated by one-way ANOVA with Holm Sidak correction for pair-wise multiple comparison. p<0.05 was considered statistically significant. Analysis was performed using Prism software (Graph Pad). Data are presented as means; error bars are standard error of the mean (SEM).
Supplementary Material
Highlights.
A fungal protease, DppIVA, cleaves and inactivates host cytokines and chemokines.
Fungal DppIVA blunts recruitment, differentiation and activation of Ly6chi monocytes.
Fungal protease cleavage of GM-CSF tempers activation of a variety of leukocytes.
An FDA approved inhibitor of DppIV ameliorates progressive fungal lung infection.
Acknowledgments
Supported by NIH R01 AI035681 (BK), AI093553 (MW) and AI106269 (GD); T32 AI055397 (SF) and GM007215 (JL); F32 AI120642 (NHS); P30ES006096 (KSV); and American Heart 15POST25700182 (KSV). Grzegorz Sabat at UW Biotechnology Center assisted with MS. Karen Ersland shared initial findings on blunted monocyte recruitment by yeast and resistance mediated by CCR2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
AS, JL, and BK conceived the experiments. AS and JL executed most experiments. TB analyzed MS data and produced rDppIVA. MW helped in study design, and infections. GD and KS helped design and perform GM-CSFR and Mϕ experiments. JL and TS generated DppIVA RNAi strains, and TB, rDppIVA. TS generated DsRed strains and AS and SF characterized them. NHS analyzed cell sources of cytokine. AS drafted the paper with BK.
REFERENCES
- Ali S, Robertson H, Wain JH, Isaacs JD, Malik G, Kirby JA. A non-glycosaminoglycan-binding variant of CC chemokine ligand 7 (monocyte chemoattractant protein-3) antagonizes chemokine-mediated inflammation. Journal of immunology. 2005;175:1257–1266. doi: 10.4049/jimmunol.175.2.1257. [DOI] [PubMed] [Google Scholar]
- Antoine-Moussiaux N, Buscher P, Desmecht D. Host-parasite interactions in trypanosomiasis: on the way to an antidisease strategy. Infection and immunity. 2009;77:1276–1284. doi: 10.1128/IAI.01185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arastu-Kapur S, Ponder EL, Fonovic UP, Yeoh S, Yuan F, Fonovic M, Grainger M, Phillips CI, Powers JC, Bogyo M. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nature chemical biology. 2008;4:203–213. doi: 10.1038/nchembio.70. [DOI] [PubMed] [Google Scholar]
- Boonacker E, Van Noorden CJ. The multifunctional or moonlighting protein CD26/DPPIV. European journal of cell biology. 2003;82:53–73. doi: 10.1078/0171-9335-00302. [DOI] [PubMed] [Google Scholar]
- Brandhorst TT, Roy R, Wuthrich M, Nanjappa S, Filutowicz H, Galles K, Tonelli M, McCaslin DR, Satyshur K, Klein B. Structure and function of a fungal adhesin that binds heparin and mimics thrombospondin-1 by blocking T cell activation and effector function. PLoS pathogens. 2013;9:e1003464. doi: 10.1371/journal.ppat.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer HE, Hoggatt J, O'Leary HA, Mantel C, Chitteti BR, Cooper S, Messina-Graham S, Hangoc G, Farag S, Rohrabaugh SL, et al. Dipeptidylpeptidase 4 negatively regulates colony-stimulating factor activity and stress hematopoiesis. Nature medicine. 2012;18:1786–1796. doi: 10.1038/nm.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B, Trapnell BC. The molecular basis of pulmonary alveolar proteinosis. Clinical immunology. 2010;135:223–235. doi: 10.1016/j.clim.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A, Decalf J, Ahloulay M, Lababidi C, Mansour H, Vallet-Pichard A, Mallet V, Mottez E, Mapes J, Fontanet A, et al. Evidence for an antagonist form of the chemokine CXCL10 in patients chronically infected with HCV. J Clin Invest. 2011;121:308–317. doi: 10.1172/JCI40594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KG, Woods JP. Secreted dipeptidyl peptidase IV activity in the dimorphic fungal pathogen Histoplasma capsulatum. Infection and immunity. 2009;77:2447–2454. doi: 10.1128/IAI.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KG, Zarnowski R, Woods JP. Histoplasma capsulatum encodes a dipeptidyl peptidase active against the mammalian immunoregulatory peptide, substance P. PloS one. 2009;4:e5281. doi: 10.1371/journal.pone.0005281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa V, Jhingran A, Dutta O, Kasahara S, Donnelly R, Du P, Rosenfeld J, Leiner I, Chen CC, Ron Y, et al. Inflammatory monocytes orchestrate innate antifungal immunity in the lung. PLoS pathogens. 2014;10:e1003940. doi: 10.1371/journal.ppat.1003940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Feng Y, Ji H, Zhang H, Zheng F, Wang C, Yin Z, Pan X, Tang J. Inactivation of dipeptidyl peptidase IV attenuates the virulence of Streptococcus suis serotype 2 that causes streptococcal toxic shock syndrome. Current microbiology. 2009;59:248–255. doi: 10.1007/s00284-009-9425-8. [DOI] [PubMed] [Google Scholar]
- Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nature reviews Immunology. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- Ingmer H, Brondsted L. Proteases in bacterial pathogenesis. Research in microbiology. 2009;160:704–710. doi: 10.1016/j.resmic.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Ishibashi Y, Matsui T, Maeda S, Higashimoto Y, Yamagishi S. Advanced glycation end products evoke endothelial cell damage by stimulating soluble dipeptidyl peptidase-4 production and its interaction with mannose 6-phosphate/insulin-like growth factor II receptor. Cardiovascular diabetology. 2013;12:125. doi: 10.1186/1475-2840-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhingran A, Mar KB, Kumasaka DK, Knoblaugh SE, Ngo LY, Segal BH, Iwakura Y, Lowell CA, Hamerman JA, Lin X, et al. Tracing conidial fate and measuring host cell antifungal activity using a reporter of microbial viability in the lung. Cell reports. 2012;2:1762–1773. doi: 10.1016/j.celrep.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BS, Vergeront JM, Weeks RJ, Kumar UN, Mathai G, Varkey B, Kaufman L, Bradsher RW, Stoebig JF, Davis JP. Isolation of Blastomyces dermatitidis in soil associated with a large outbreak of blastomycosis in Wisconsin. N Engl J Med. 1986;314:529–534. doi: 10.1056/NEJM198602273140901. [DOI] [PubMed] [Google Scholar]
- Krajaejun T, Gauthier GM, Rappleye CA, Sullivan TD, Klein BS. Development and application of a green fluorescent protein sentinel system for identification of RNA interference in Blastomyces dermatitidis illuminates the role of septin in morphogenesis and sporulation. Eukaryot Cell. 2007;6:1299–1309. doi: 10.1128/EC.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y, Konishi K, Gomi T, Yagishita H, Yajima A, Yoshikawa M. Enzymatic properties of dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis and its participation in virulence. Infection and immunity. 2000;68:716–724. doi: 10.1128/iai.68.2.716-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison BE, Park SJ, Mooney JM, Mehrad B. Chemokine-mediated recruitment of NK cells is a critical host defense mechanism in invasive aspergillosis. The Journal of clinical investigation. 2003;112:1862–1870. doi: 10.1172/JCI18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X, O'Leary HA, Broxmeyer HE. Implications of DPP4 modification of proteins that regulate stem/progenitor and more mature cell types. Blood. 2013;122:161–169. doi: 10.1182/blood-2013-02-487470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Critical reviews in microbiology. 2010;36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- Qi SY, Riviere PJ, Trojnar J, Junien JL, Akinsanya KO. Cloning and characterization of dipeptidyl peptidase 10, a new member of an emerging subgroup of serine proteases. The Biochemical journal. 2003;373:179–189. doi: 10.1042/BJ20021914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Meng XP, Ramasamy S, Harrison DG, Galis ZS. Reactive oxygen species produced by macrophage-derived foam cells regulate the activity of vascular matrix metalloproteinases in vitro. Implications for atherosclerotic plaque stability. J Clin Invest. 1996;98:2572–2579. doi: 10.1172/JCI119076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci U S A. 2007;104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Goldman WE. Fungal stealth technology. Trends in immunology. 2008;29:18–24. doi: 10.1016/j.it.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annual review of immunology. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Sullivan TD, Rooney PJ, Klein BS. Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot Cell. 2002;1:895–905. doi: 10.1128/EC.1.6.895-905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touz MC, Nores MJ, Slavin I, Piacenza L, Acosta D, Carmona C, Lujan HD. Membrane-associated dipeptidyl peptidase IV is involved in encystation-specific gene expression during Giardia differentiation. The Biochemical journal. 2002;364:703–710. doi: 10.1042/BJ20020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JI, Weeks M, Allen D, Hutcheson RH, Jr, Anderson R, Fraser DW, Kaufman L, Ajello L, Spickard A. Acute histoplasmosis: clinical, epidemiologic and serologic findings of an outbreak associated with exposure to a fallen tree. The American journal of medicine. 1979;66:587–595. doi: 10.1016/0002-9343(79)91168-9. [DOI] [PubMed] [Google Scholar]
- Wuthrich M, Ersland K, Sullivan T, Galles K, Klein BS. Fungi subvert vaccine T cell priming at the respiratory mucosa by preventing chemokine-induced influx of inflammatory monocytes. Immunity. 2012;36:680–692. doi: 10.1016/j.immuni.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazbeck R, Howarth GS, Abbott CA. Dipeptidyl peptidase inhibitors, an emerging drug class for inflammatory disease? Trends in pharmacological sciences. 2009;30:600–607. doi: 10.1016/j.tips.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Youseff BH, Holbrook ED, Smolnycki KA, Rappleye CA. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS pathogens. 2012;8:e1002713. doi: 10.1371/journal.ppat.1002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.