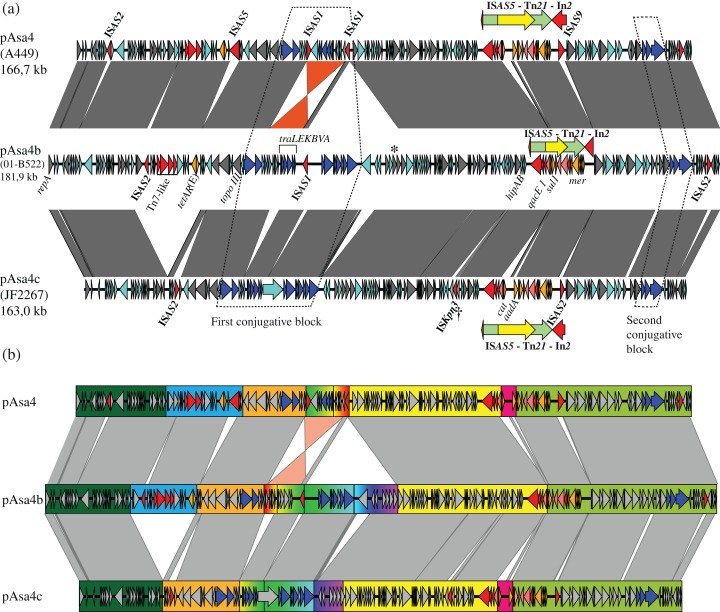
Figure 1. Nucleotide alignment of three plasmid variants: pAsa4, pAsa4b, and pAsa4c.
(A) Plasmid alignments and ORF representations were done using EasyFig (Sullivan, Petty & Beatson, 2011). The dark grey bands denote regions of identity. Overall, the identity was more than 94%. The bands of non-contiguous repeat regions were removed for clarity. An inversion between pAsa4 and pAsa4b/c is marked in orange. ORFs are indicated by colored arrows that indicate their deduced function: Cyan: hypothetical protein; Dark grey: maintenance and replication; Blue: type IV secretion system-like conjugative system, contained in two conjugative blocks (dashed lines); Red: IS, transposition-associated genes; Orange: antimicrobial resistance. The following features have also been annotated: Asterisk: phage endonuclease, similar to pAsa4 pseudogene; Dagger: IS1595-family ISKpn3. A transposon/integron structure (Tn21/In2) that was common to all pAsa4 plasmids and that is integrated into an ISAS5 is indicated over each plasmid by nested red, green, and yellow arrows. Besides the transposon, but inside ISAS5, an ISAS9 and an ISAS2 insertion could be seen for pAsa4 and pAsa4c, respectively. (B) Segments of large insertion/deletion or recombination sequences are highlighted in color. Segments of particular significance are: Blue: an insertion/deletion in pAsa4 comprising tetracycline resistance genes tetAR (E); Rainbow progression: multiple insertions/deletions and an inversion encompassing a conjugative gene region; Pink: two events: an ISCR insertion comprising the chloramphenicol resistance gene, cat and an aadA1 cassette.