Figure 2.
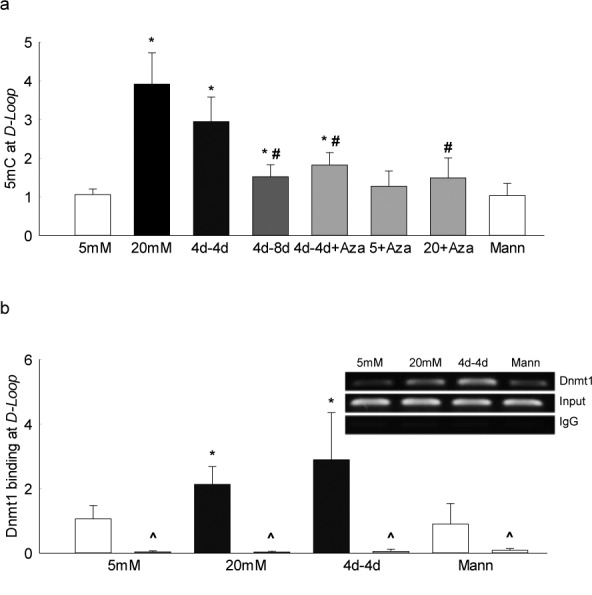
Effect of reversal of high glucose on mtDNA methylation. DNA methylation of the D-loop region of the mtDNA was quantified by (a) 5mC levels using methylated DNA immunoprecipitation technique, and (b) by Dnmt1 binding in the crosslinked retinal endothelial cells using Dnmt1 antibody, followed by amplification of the D-loop region by q-PCR; IgG was used as an antibody control (marked as ^). Fold change was calculated relative to the values obtained from cells in continuous 5 mM glucose. Data are represented as mean ± SD from each measurement made in duplicate in three to four cell preparations. 4d-4d and 4d-4d+Aza = 4 days of 20 mM glucose, followed by 4 days of 5 mM glucose in the absence or presence of Aza, respectively; 4d-8d = cells in 5 mM glucose for 4 days followed by 20 mM glucose for 8 days; 5+Aza and 20+Aza = cells in continuous 5 mM glucose or 20 mM glucose, in the presence of Aza. *P < 0.05 vs. 5 mM glucose; #P < 0.05 vs. 20 mM glucose.
