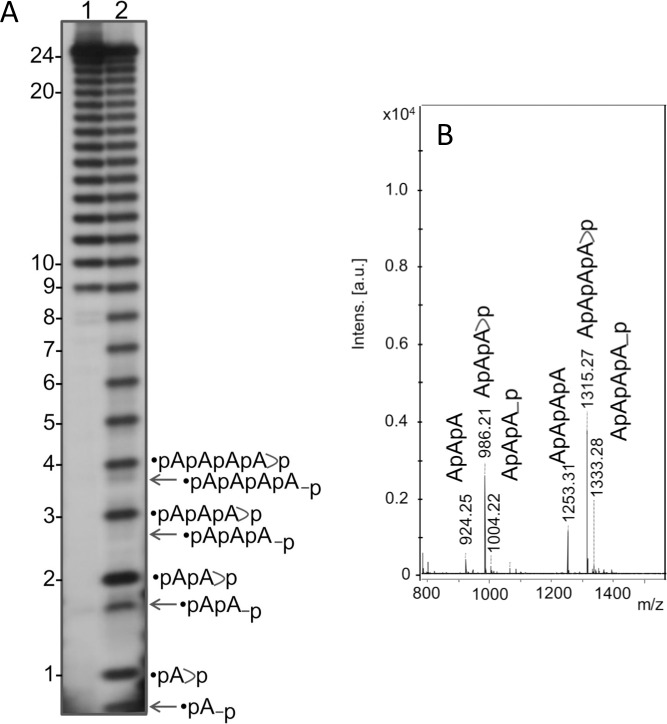
Fig 4. Hydrolysis of A24 by water or formamide.
Panel A: 20% acrylamide PAGE analysis of 5’ labeled A24, hydrolyzed in water (Lane 1, see text) or formamide (Lane 2, see text). The product formed from A24 (not phosphorylated at 5’) upon digestion in formamide was analyzed by MALDI in the negative ion mode. Panel B shows, as examples, the MALDI profiles corresponding to the trimer and tetramer families produced by formamide hydrolysis. The hydrolysis produces: (i) a species with m/z = 986.21 corresponding to the primary product of the cleavage at 3’ = ApApA>p, (ii) a species with m/z = 1004.2 (986 + 18) derived from its opening to ApApA_p, (iii) a species with m/z = 924.25 (1004–80) derived by the phosphate-loss of this latter = ApApA. Analogous products can be derived for the tetramer: 1315.27 (ApApApA>p) opened to 1333.28 (ApApApA_p) and dephosphorylated to 1253.31 (ApApApA). The fully dephosphorylated species migrates in the gel faster than the phoshorylated ones. Likewise, in MALDI this species gives a signal at lower m/z value. Longer oligomers behave similarly. Thus, using a reference ladder produced this way for the interpretation of the 3’,5’ cAMP oligomerization products is fully justified. Note that in contrast to the oligomers detected by MALDI, the oligomers detected by PAGE always carry a 32P-phosphate group at 5’.