Figure 2. Crystal Structure of MBP-Casp-8tDED-F122G/L123G and Cryo-EM Reconstruction of the GFP-Casp-8tDED Filament.
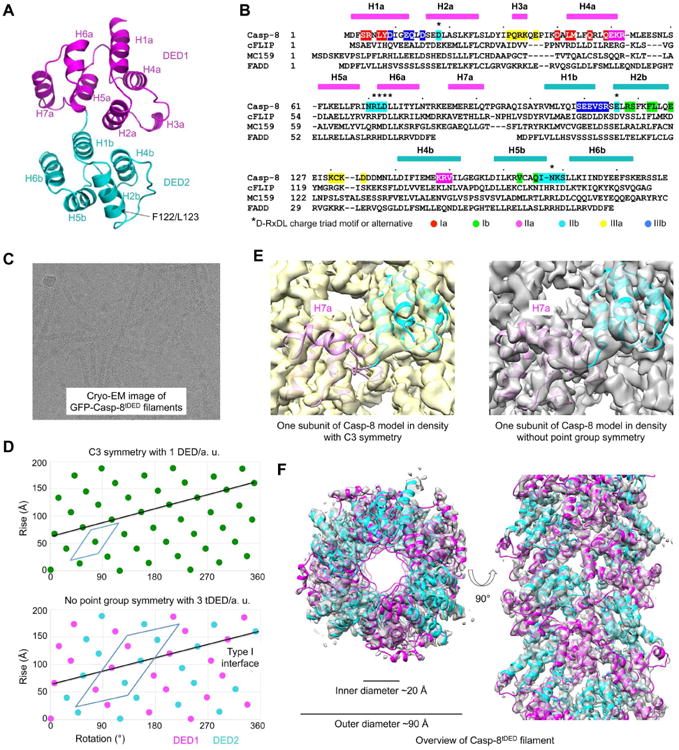
(A) A ribbon diagram of the Casp-8tDED (F122G/L123G) crystal structure labeled with secondary structures, and locations of the mutations.
(B) Sequence alignment among Casp-8, cFLIP, MC159 and FADD. Secondary structures are labeled on the top. Key residues involved in filament assembly are highlighted with type Ia in red, type Ib in green, type IIa in magenta, type IIb in cyan, type IIIa in yellow and type IIIb in blue.
(C) A cryo-electron micrograph of GFP-Casp-8 tDED filaments.
(D) Helical net plots showing the asymmetric units of the filament in C3 symmetry (top) and without point group symmetry (bottom), and the direction of the type I interaction that connects DED1 and DED2 in tDED.
(E) One subunit of the Casp-8tDED model fitted into cryo-EM densities reconstructed in C3 symmetry (left) and without point group symmetry (right). The density for H7a is absent in the C3 map.
(F) Top and side views of the GFP-Casp-8tDED filament structure superimposed with the final cryo-EM density.
See also Figure S2.
