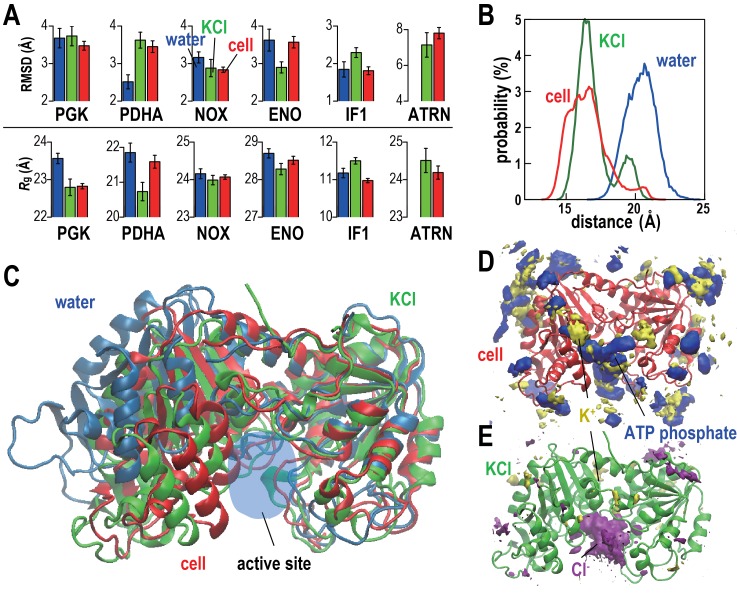
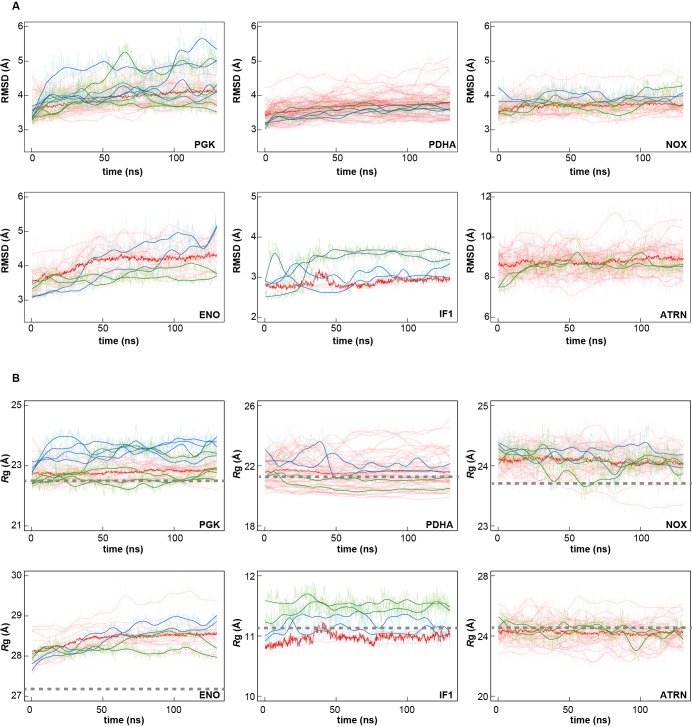
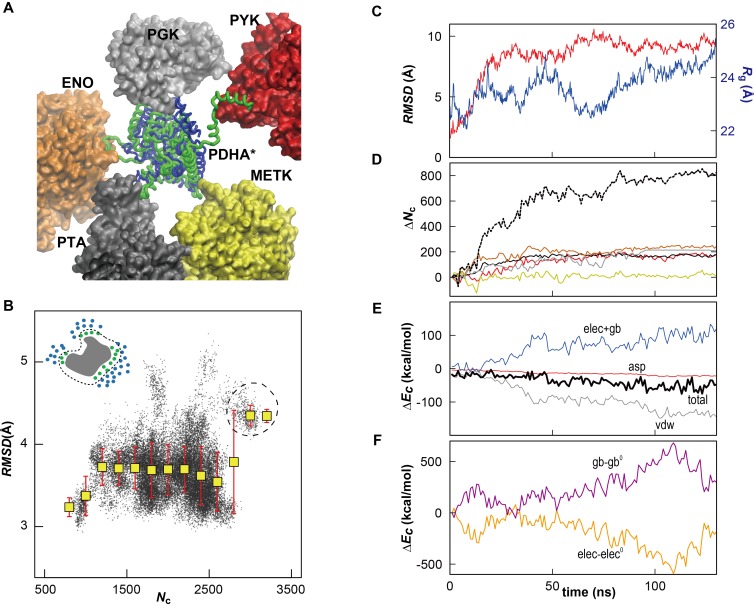
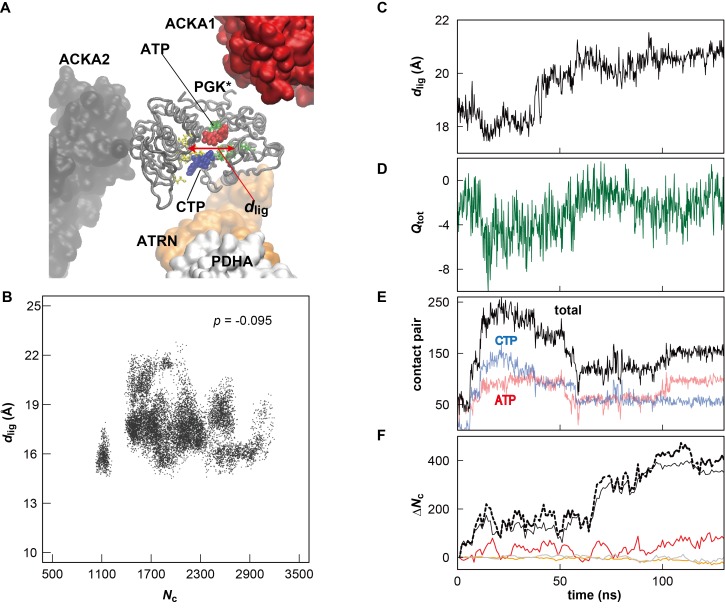
(
A) Denatured (green) conformation of one of the 39 copies of PDHA (denoted as PDHA*) due to contacts with other cytoplasmic proteins (PYK (red), PGK (gray), ENO (orange), PTA (black), and METK (yellow)). The initial, native, homology model is shown in blue. (
B) Correlation between coordination number of crowder C
α atoms
Nc (see Materials and methods) and
RMSD (based on C
α atoms relative to the initial model) for PDHA. The schematic figure in upper left shows the target protein (gray) surrounded by cytoplasmic proteins (blue). The atoms counted in
Nc (green) are shown in green. Histogram averages are shown as yellow boxes with standard deviations indicated as red bars. The dashed circle corresponds to the denatured state shown in panel A. Instantaneous values of
Nc,
dlig and RMSD were calculated using an interval of 200 ps using 39 copies of PDHA, respectively, in the
MGm1 system. (
C) Time history of
RMSD (based on C
α atoms relative to the structure after the equilibration) (red) and radius of gyration (
Rg) (blue) of PDHA*. (
D) Time history of the contact pair between all atoms in the PDHA* and all atoms in five vicinal proteins (line color corresponds to those proteins in panel A) with the total value of them (dashed black). The schematic figure in the upper left shows the contact pairs (dashed line) between the atoms in two proteins (green and blue). The cutoff distance of the contact pair was set to 10 Å. (
E) Time histories of the energy changes of PDHA* upon crowding (
ΔEc) (i.e. the energy of all proteins (PDHA* with 5 vicinal proteins) subtracted by those energies of isolated PDHA* and the five vicinal proteins). Each energy component of
ΔEc is shown with different colors (van der Waals energy (vdw; gray), cost of cavity formation calculated as 0.005 cal/mol/Å
2 * SASA (asp; red), combination of the vacuum electrostatic and electrostatic solvation energies (elec+gb; blue), and the total energy (tot; thick black line). (
F) Time history of the electrostatic Coulomb energy (elec; orange) and electrostatic solvation energy obtained via the GBMV generalized Born method in CHARMM (
Lee et al., 2003) (gb; violet) where energies of PDHA* upon crowding relative to the initial values were elec
0 = −1399 and gb
0 = 1406 kcal/mol, respectively.