Figure 4. Characterization of the Ae. aegypti methylome by whole-genome bisulfite sequencing.
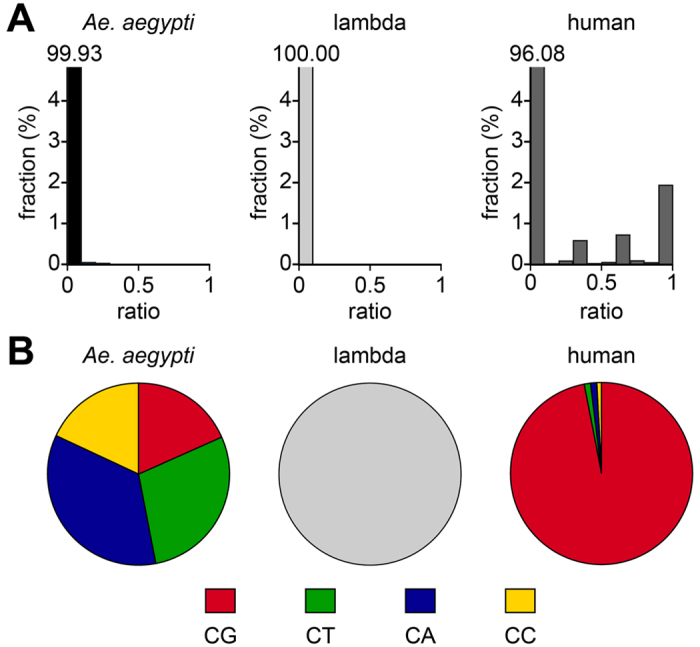
(A) Average methylation levels were determined for all cytosine residues and then distributed into bins with increasing methylation ratios. For comparison, the corresponding data is also shown for bacteriophage lambda (negative control) and human blood (positive control) DNA that was spiked into the Ae. aegypti sample prior to bisulfite conversion. The actual numerical values of the first bins are indicated above the corresponding bars. (B) Dinucleotide sequence context of unconverted cytosines. No unconverted cytosines were found in the lambda dataset.
