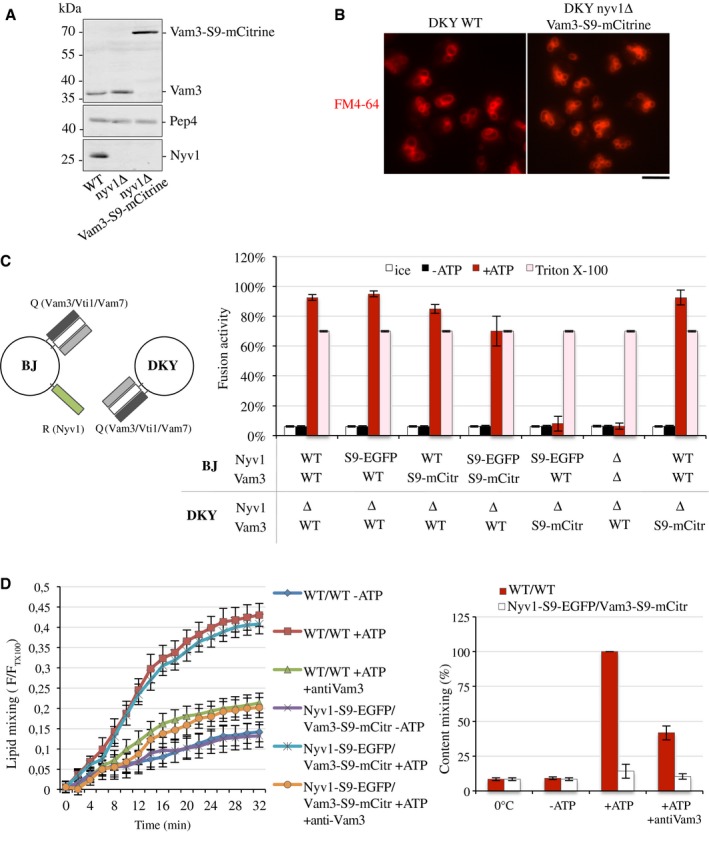
Vacuoles were isolated from BJ3505 and DKY6281 cells with SNAREs tagged or deleted as indicated. Note that the presence of Nyv1 on only one fusion partner still permits efficient fusion
64 but ensures that
trans‐SNARE complexes can only form in one orientation, between the Q‐SNARE in DKY6281 and the R‐SNARE in BJ3505. The vacuoles were used in standard fusion reactions, and content mixing was measured via the alkaline phosphatase assay. In parallel, identical samples were incubated either on ice, which prevents fusion, or in the presence of 0.5% Triton X‐100, which allows fusion‐independent access of the maturase Pep4 to pro‐ALP and controls for the levels of these two reporter enzymes in the samples. Means ± s.d. are shown from three independent experiments.