Figure 5. CDC2b from A. castellanii could be the regulator of the cell cycle.
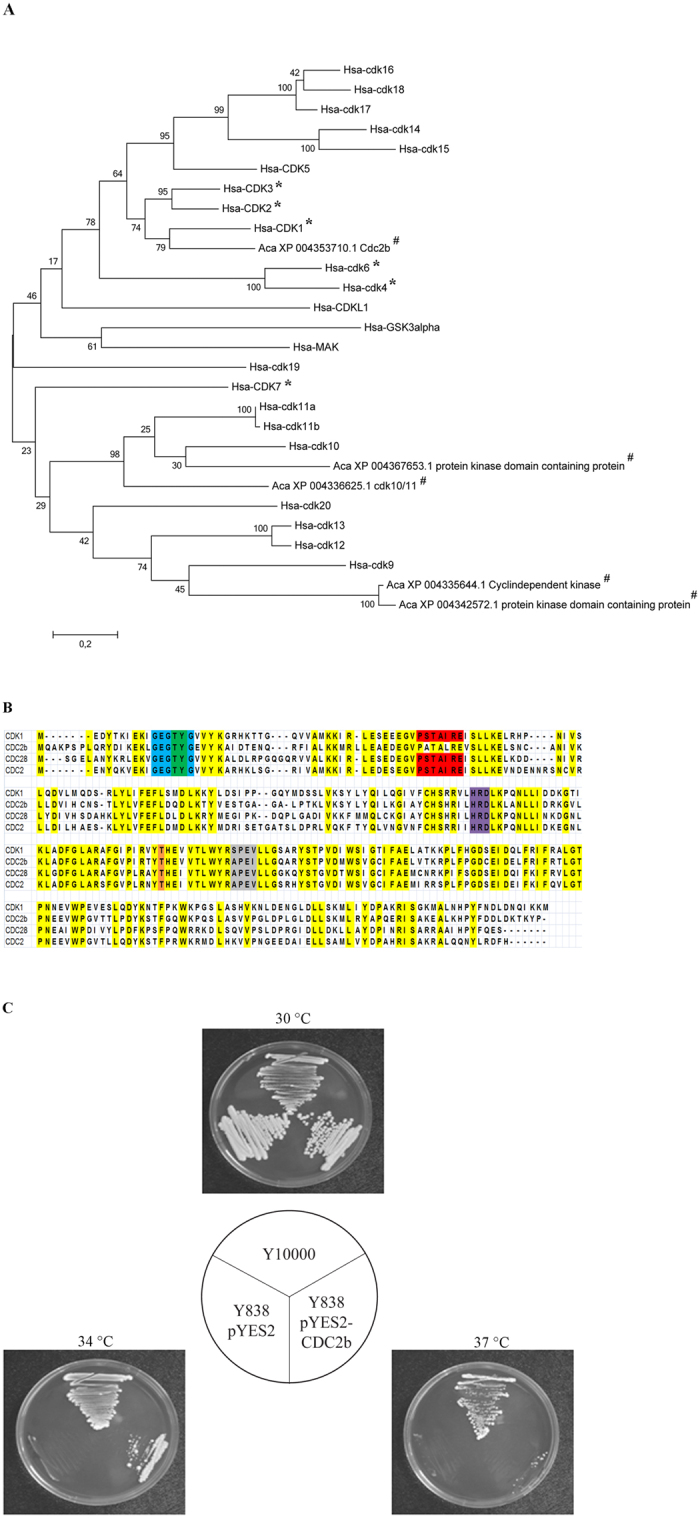
(A) CDK phylogeny inferred by Maximum Likelihood based on human (Hsa) CDKs18 and putative A. castellanii (Aca) CDKs. 1000 bootstrap replications were inferred and displayed at the nodes. The scale bar represents the number of amino acid substitution per site. All proteins were first labeled with their species name (Aca, A. castellanii; Hsa, Homo sapiens). *Indicates humans cell cycle-related proteins and # represents proteins from A. castellanii. (B) Sequence alignment between CDC2b (XP_004353710.1) from A. castellanii, CDK1 (NP_001777.1) from Homo sapiens, CDC28 (NP_009718.3) from Saccharomyces cerevisiae and CDC2 (CAC37513.1) from Schizosaccharomyces pombe using MUSCLE of the MEGA6 software. Yellow color highlight conserved residues. Motifs characteristics of CDK are displayed in blue (ATP-binding domain), green (inhibitory phosphorylation sites), red (cyclin-binding domain), purple (start of T-loop), orange (activating phosphorylation site) and grey (end of T-loop). (C) The S. cerevisiae strains Y10000 (wild type), Y838 (cdc28-4) transformed with the empty plasmid pYES2 or the recombinant plasmid pYES2-CDC2b were streaked on galactose-containing medium and cultivated at 30 °C, 34 °C or 37 °C for 4 days.
