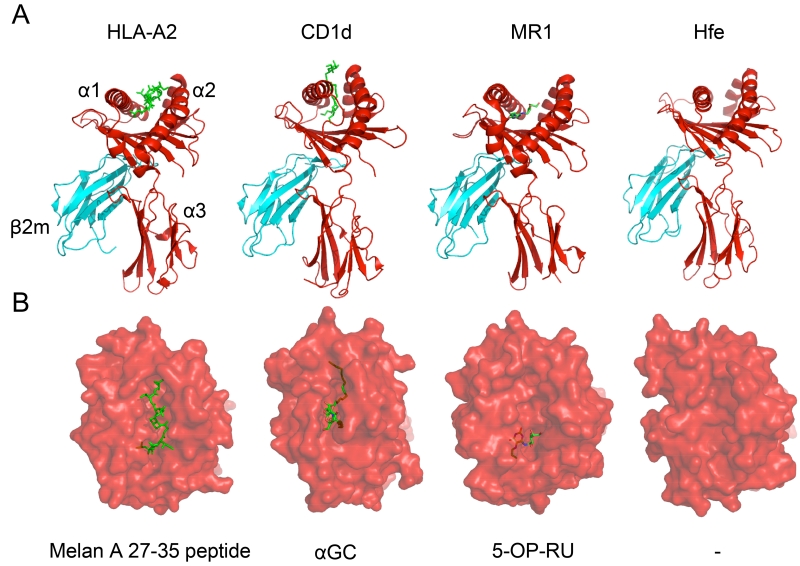
Figure 2. Structural Views of Ligands Bound to Different MHC-Ia and MHC-Ib Proteins.
In order to demonstrate the similarities and differences between MHC-Ia and MHC-Ib molecules, the HLA-A2 (PDB: 2GTW), hCD1d (PDB: 3SDX), hMR1 (PDB: 4PIC), and hHFE (PDB: 1A6Z) proteins are provided as examples. In panel A, the heavy chain is displayed in red with its three structural domains also denoted. The monomorphic β2m molecule, which associates with most MHC-Ib molecules, is portrayed in cyan as it interacts with the α3 domain of the MHC molecules. Finally, the antigen for each protein (except for hHFE) is illustrated in neon green. All proteins adopt a similar fold and use two α-helices to form the ligand-binding pocket while anti-parallel β-strands form the floor of the pocket. The pocket for hCD1d is much deeper than the ones present in HLA-A2 and hMR1, distinguishing the antigens each molecule presents. In panel B, a top-down illustration is used to represent the view of the antigen-MHC complex from the TCR’s perspective. HLA-A2 is shown presenting a melanoma-specific protein while hCD1d and hMR1 are presenting their prototypic stimulatory antigens α-galactosylceramide (α-GC) and 5-OP-RU, respectively. hHFE possesses too narrow and flat a groove to appropriately present an antigen. In this representation, it is easy to visualize that while HLA-A2 presents an antigen that spans the length of its groove, hMR1 presents an antigen that is sequestered exclusively in the A’ pocket.