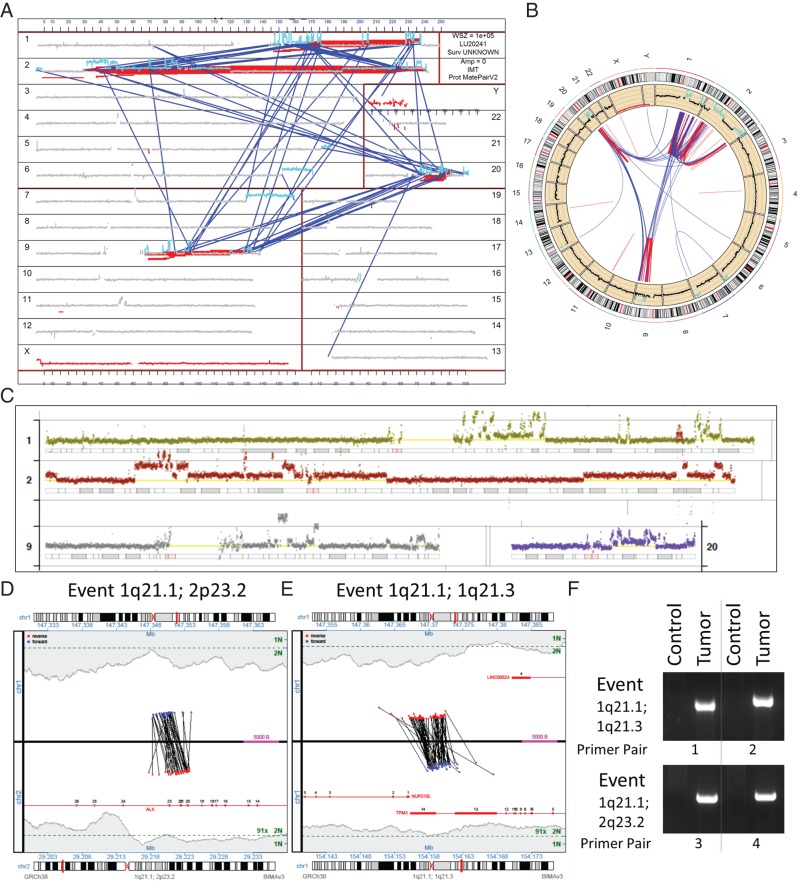
Figure 2.
Molecular characterization of treatment-resistant tumor sample. The multiple rearrangements are demonstrated in a genome plot of all structural variations in the tumor (A) and a conventional circos plot (B). The genome plot presents coverage (y-axis) across each chromosome (x-axis) with a window size (WSZ) of 1 × 105. Gray indicates normal diploid coverage, with gains indicated in cyan. Inter- and intrachromosomal rearrangements are presented as blue and red lines linking chromosome positions, analogous to the circos plot. The q-arms of chromosomes 1, 9 and 20, together with multiple regions of chromosome 2 including one containing the ALK locus were involved in a chromoplectic rearrangement, with more detailed coverage across each chromosome presented in (C). The normal diploid two-copy level is presented by the yellow lines for each chromosome. Junction plots present the mate pair reads supporting the translocation between intron 19 of ALK at 2p23.2 (lower panel of plot) and chromosome 1 (1q21.1) at a non-genic region between CHD1L and LINC00624 (upper panel of plot) (D). A second junction plot describes an intra-chromosomal rearrangement adjacent to this chromosome one position at 1q21.1, linking to the final intron of TPM3 at 1p21.1 (E). Red and blue dots represent mate pair reads mapping to the positive or negative DNA strands, respectively, with coverage levels across each position presented in gray-shaded regions, with single (1N) and two-copy levels (2N) indicated (green). Gene regions are presented in red with exons numerated. Both the ALK (1q21.1; 2p23.2) and TPM3 (1p21.1; 1p21.3) events were verified by PCR on tumor DNA using different primer combinations spanning the breakpoint junctions (F).