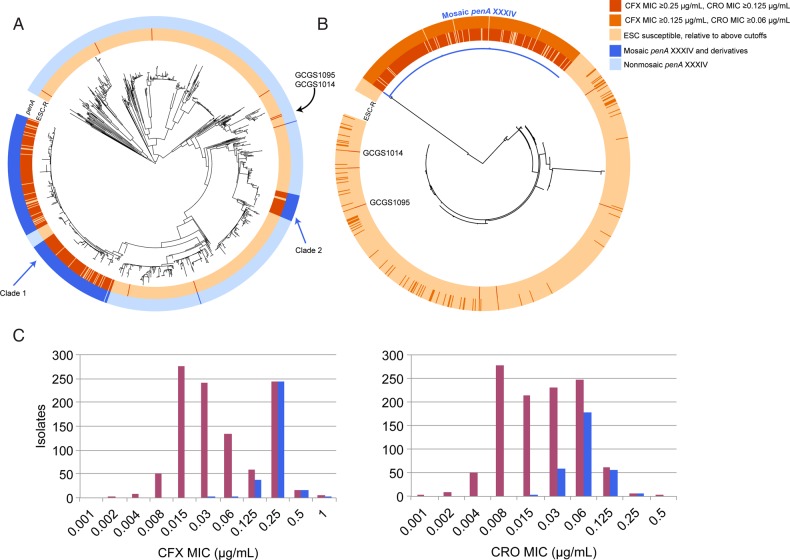
Figure 2.
A, Maximum likelihood whole-genome-sequence phylogeny of 1102 Neisseria gonorrhoeae isolates, based on single-nucleotide polymorphisms from mapping to the FA1090 reference genome, with the inner annotation ring representing those isolates with reduced extended-spectrum cephalosporin (ESC) susceptibility and the outer ring representing which isolates have a mosaic penA XXXIV or derivative allele. Clades 1 and 2 are identified. A lineage within clade 1 lacks the mosaic penA XXXIV and is susceptible; these isolates appear to have undergone a recombination that replaced the mosaic penA XXXIV allele with mosaic penA XXXVIII allele. B, Maximum likelihood phylogeny of the penA locus, extracted from the de novo–assembled genomes for each of the isolates. The branch in blue indicates isolates with the mosaic penA XXXIV and derivative alleles. The coloring along the inner and outer annotation rings indicate isolates by the minimum inhibitory concentration (MIC) threshold defined in the key. C, Histograms indicating the number of isolates by cefixime (CFX) and ceftriaxone (CRO) MICs. The histogram in purple indicates the total number of isolates—those with and without a mosaic penA XXXIV-like allele—per MIC, and the histogram in blue indicates the number of isolates with a mosaic penA XXXIV–like allele.