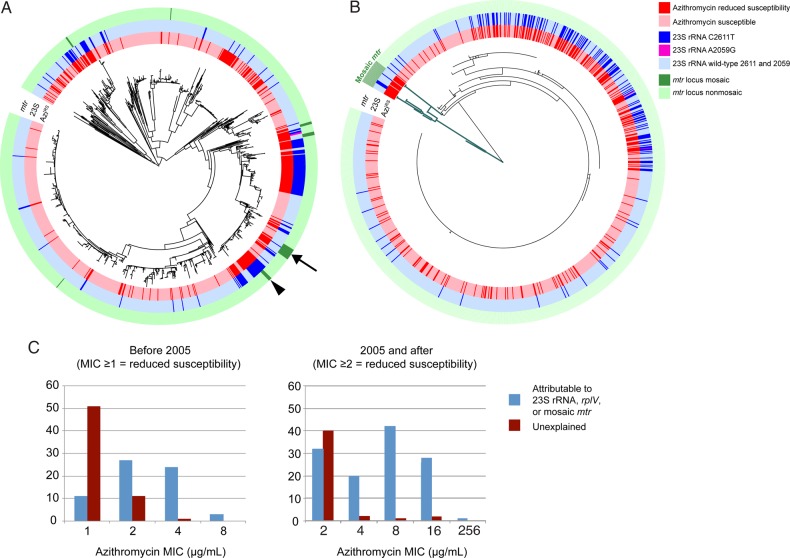
Figure 4.
A, Maximum likelihood whole-genome-sequence phylogeny of 1102 Neisseria gonorrhoeae isolates, based on single-nucleotide polymorphisms from mapping to the FA1090 reference genome, with the inner annotation ring representing reduced azithromycin susceptibility, the middle annotation ring indicating isolates with at least 2 copies of the C2611T 23S ribosomal RNA (rRNA) mutation and 2 isolates with 4 copies of the A2059G 23S rRNA mutation, and the outer annotation ring indicating isolates with a mosaic mtr locus. The arrow indicates an example where the mtr locus mosaic is inferred to have appeared first, followed by acquisition of the C2611T mutation, and the wedge indicates an example with the opposite order of acquisition. B, Maximum likelihood phylogeny of the mtrR locus including the 200 base pairs upstream of the coding sequence start site, extracted from the de novo–assembled genomes for each of the isolates. The branches in green indicate isolates with mosaic mtr loci. As in panel A, the annotation rings proceed from the innermost being reduced azithromycin susceptibility to the outermost being the mosaic mtr loci. C, Histograms indicating the azithromycin minimum inhibitory concentrations (MICs), separated into 2 sections by when there was a change in the azithromycin MIC testing protocol, such that the cutoff changed from 1 to 2 µg/mL.