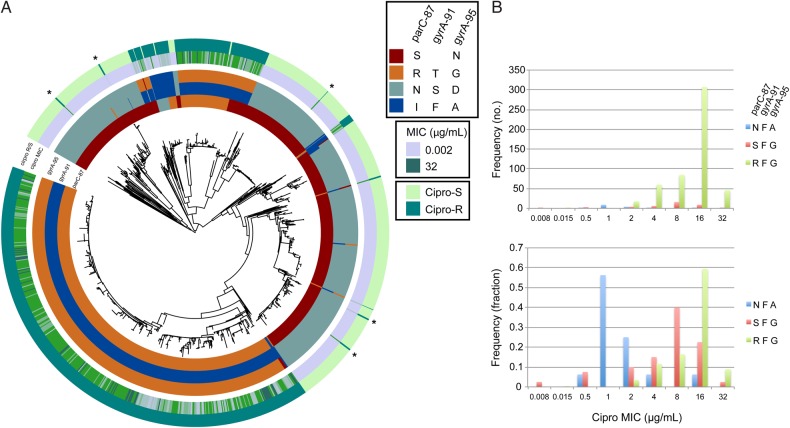
Figure 5.
A, Maximum likelihood whole-genome-sequence phylogeny of 1102 Neisseria gonorrhoeae isolates, based on single-nucleotide polymorphisms from mapping to the FA1090 reference genome, with the inner annotation rings representing amino acid residues at ParC-87, GyrA-91, and GyrA-95. The outer annotation rings represent ciprofloxacin (Cipro) minimum inhibitory concentrations (MICs) and dichotomized resistance (R) and susceptibility (S; cutoff for resistance at 1 µg/mL). The asterisks indicate quinolone-resistant N. gonorrhoeae that lack the ParC-87, GyrA-91, and GyrA-95 variants. B, Histograms of Cipro MICs of isolates based on haplotypes at ParC-87, GyrA-91, and GyrA-95 in absolute frequency (upper histogram) and in fraction (lower histogram).