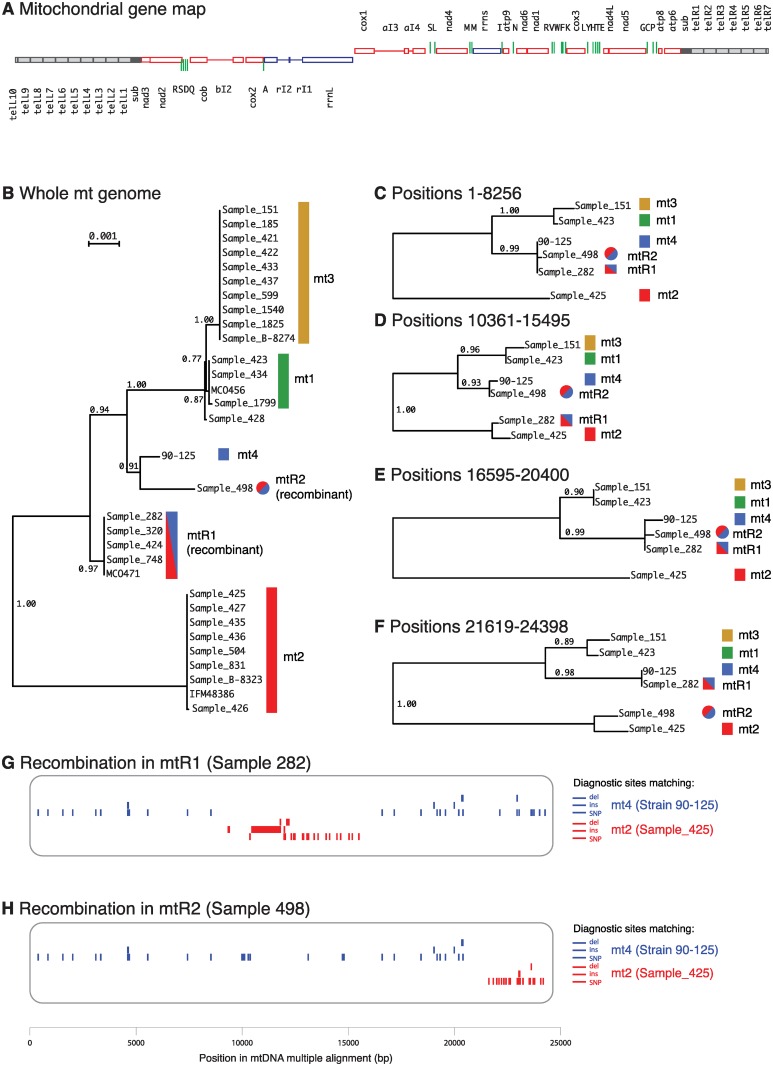
Fig 6. Phylogeny and recombination of C. orthopsilosis mitochondrial genomes.
A. Gene map of the PacBio assembly of the mitochondrial genome from Sample 427 (33,782 bp). Subterminal repeats are black and telomeric repeats are grey. All genes from cox1 to the right telomere are transcribed rightwards, and all genes from rrnL to the left telomere are transcribed leftwards. Thin horizontal lines represent introns. Green vertical lines are tRNA genes. B. Phylogenetic tree of the whole mitochondrial genome (excluding subterminal and telomeric regions) from all C. orthopsilosis strains, generated by PhyML from a Clustal Omega alignment in Seaview. (C–F). Phylogenetic trees of four sections of the genome, for strains representing mitochondrial haplotypes mt1-mt4 and recombinants. (G, H) Inference of recombination sites in recombinant mtDNAs. For each mitochondrial haplotype mt1-mt4 we identified SNPs, insertions and deletions that are unique to that haplotype (not shared with the other three). We then scanned the putatively recombinant strains Sample 282 (mtR1) and Sample 498 (mtR2) for the presence of these unique sites. Neither of these strains shares any unique sites with mt1 or mt3. We then repeated the analysis, defining a larger set of sites that are diagnostic for mt2 (not shared with mt4) or diagnostic for mt4 (not shared with mt2).