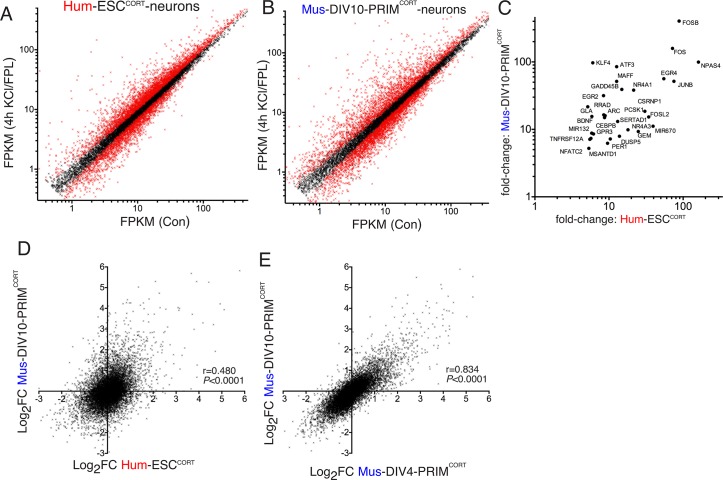
Figure 1. Conservation and divergence in gene regulation in neurons of human and mouse origin.
(A,B) Analysis of gene expression changes induced by KCl/FPL in Hum-ESCCORT-neurons (A) and DIV10 Mus-PRIMCORT-neurons (B). Normalised RNA-seq read density (FPKM) mapping to each gene in RNA extracted from control vs. KCl/FPL-treated neurons is shown (n = 3 independent biological replicates). Genes whose expression was significantly altered by KCl/FPL treatment (Benjamini-Hochberg-adjusted p-value<0.05, calculated within DESeq2) are highlighted in red. (C) Cohort of human:mouse orthologous pairs where both are strongly (>5-fold) and significantly (Benjamini-Hochberg-adjusted p-value<0.05) induced in Hum-ESCCORT-neurons and DIV10 Mus-PRIMCORT-neurons respectively. (D) Correlation of KCl/FPL-induced fold-change in 11,302 ortholog pairs in DIV10 Mus-PRIMCORT-neurons vs. DIV10-Hum-ESCCORT-neurons. (E) Correlation of KCl/FPL-induced fold-change in the same 11,302 genes as in (D) in DIV10 vs. DIV4 Mus-PRIMCORT-neurons.
DOI: http://dx.doi.org/10.7554/eLife.20337.003