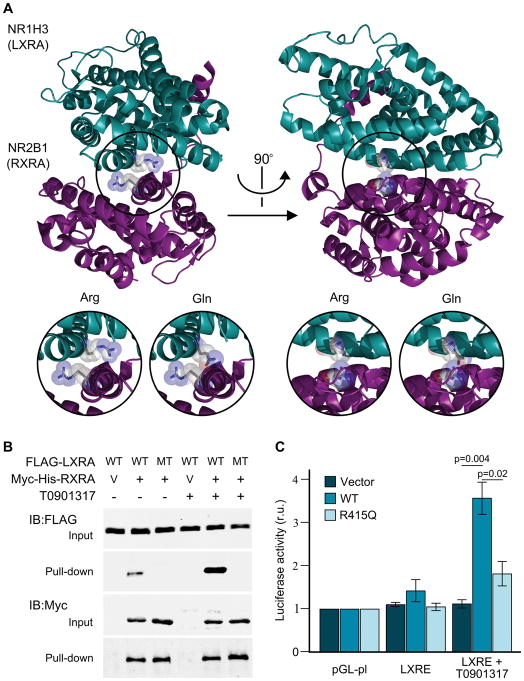
Figure 3.
Structural and functional analysis of NR1H3. A) Crystal structure of NR1H3 (LXRA) and NR2B1 (RXRA) heterodimer showing the highly conserved arginine residues in their corresponding ligand binding domain was generated with PyMol (PDB ID: 2ACL). Inserts provide a detailed view of the LXRA wild-type (Arg) and mutated (Gln) protein structure. B) FLAG-LXRA wild type (WT) and p.Arg415Gln mutant (R415Q) were co-expressed with RXRA-mycHis (+) or empty vector pcDNA4-myc-his-A (V) in HEK cells with or without 10μM LXRA agonist T0901317 overnight. RXRA was pulled down from the lysates with nickel-charged magnetic beads. Cell lysates and precipitates were immunoblotted with FLAG or Myc antibodies. C) Transcriptional activation of control (pGL-pl) and LXR response element-containing plasmids (LXRE) after co-transfection with empty vector, wild-type (WT) or mutant LXRA (R415Q). Firefly/Renilla luciferase activity (mean ± standard error) for the different constructs, with and without the presence of 10μM of a LXRA agonist (T0901317) normalized to pGL-pl under the same conditions are provided. Statistically significant Fisher’s Least Significant Difference (LSD) post hoc values are provided. r.u., relative units.