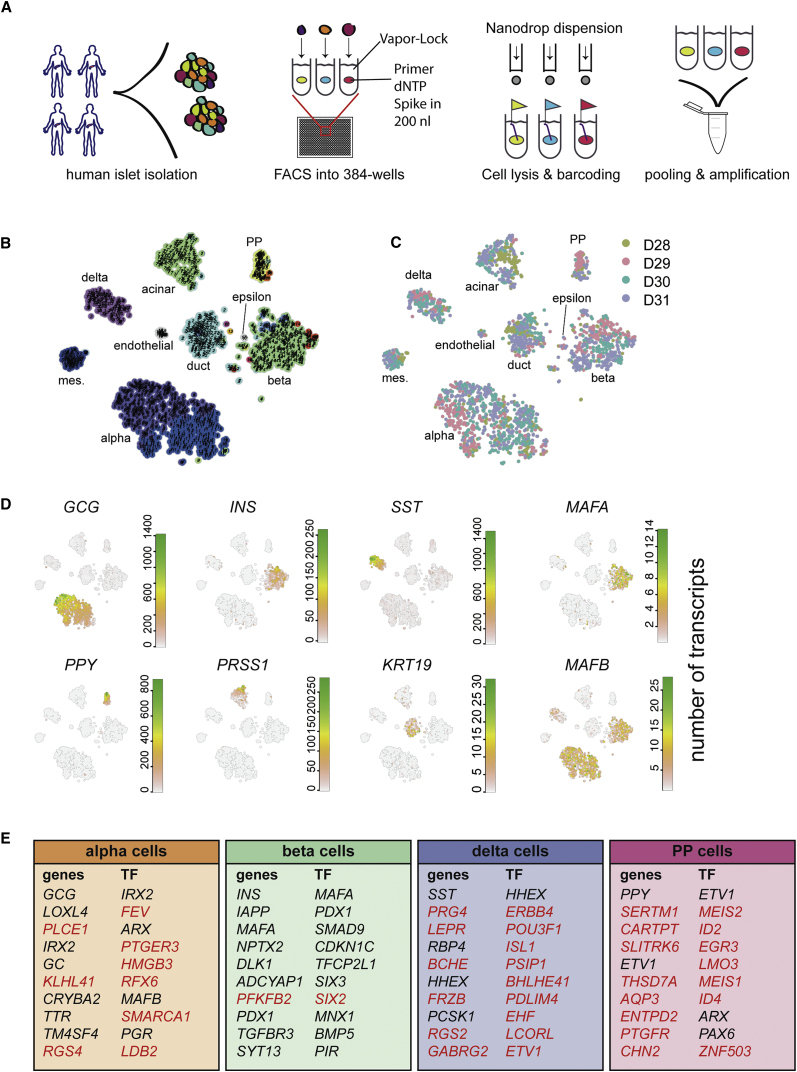
Figure 1.
SORT-Seq Allows for Deep Sequencing of Human Pancreas Cells
(A) Experimental workflow for SORT-seq. Islets were isolated from human donors. Cells were dispersed and sorted into 384-well plates with mineral oil, containing 100 nL of CEL-seq2 primers, dNTPs, and spike-ins. The RT mix was then distributed by Nanodrop II. After second-strand synthesis, material was pooled and amplified before RNA library preparation.
(B) Visualization of k-medoid clustering and cell-to-cell distances using t-SNEs. Each dot represents a single cell. Colors and numbers indicate clusters, and cell-type names are indicated with their corresponding cluster or clusters.
(C) t-SNE map highlighting donor source. Each color represents one donor.
(D) t-SNE maps highlighting the expression of marker genes for each of the six main pancreatic cell types. Transcript counts are given in a linear scale.
(E) Tables denoting the top ten differentially expressed genes and transcription factors (TFs) when comparing one cell type to all other cells in the dataset (p < 10−6). Genes whose cell-type specificity was previously unknown in the human pancreas are marked in red.