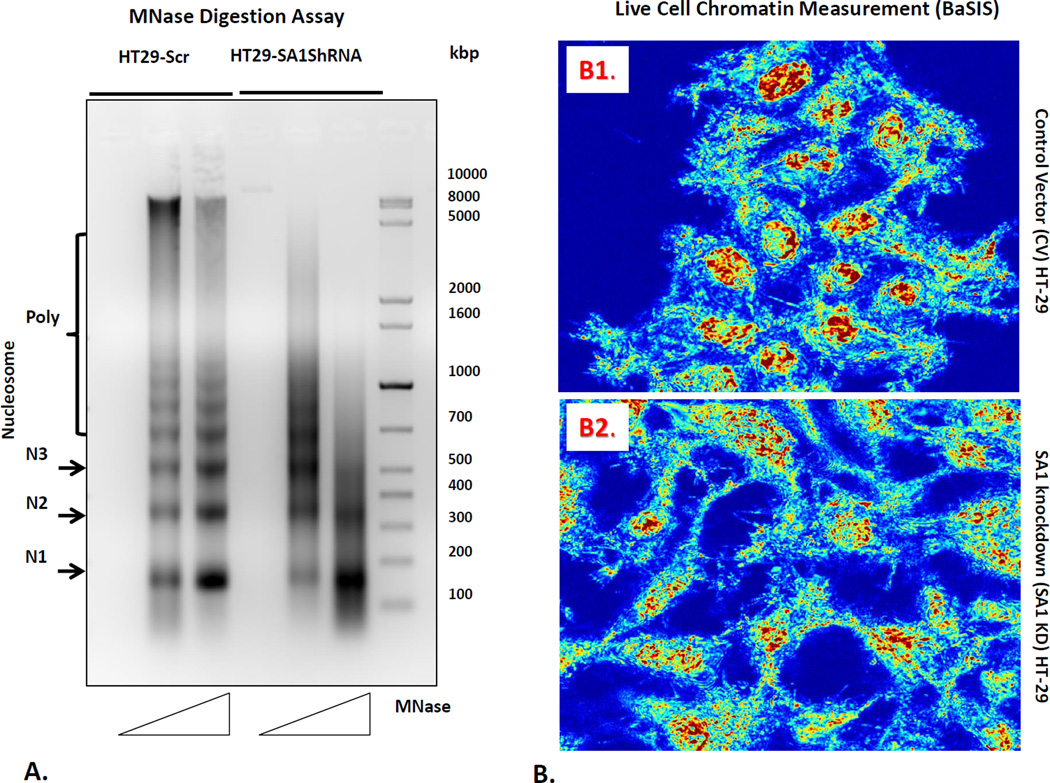
Figure 4.
Chromatin modifications related to SA1 downregulation. For these studies, we first performed (panel A) micrococcal nuclease digestion assay to determine the nucleosome positioning pattern of the nuclei isolated from HT 29 cells and SA1 shRNA transfected HT29 cells. Nuclei were digested with increasing concentrations of the MNase enzyme (50 and 100 units) for 5min followed by DNA extraction using phenol-chloroform. 5µg of DNA was then separated on a 1.5% agarose gel. Mono (N1), -di (N2), -tri (N3) and poly-nucleosomes are indicated on the left. Furthermore, we measured nanoscale chromatin alterations (panel B) in live cells using the novel technique, Back-Scattering Interference Spectroscopic (BaSIS) microscopy. BaSIS quantifies the spatial fluctuations in the z direction, measured as Σ, with sensitivity to structures between 20–200nm that can quantify the dynamics of the nano-molecular organization in live cells without using exogenous labels. Control (Panel B1) and SA-1 shRNA knockdown (Panel B2) cells were imaged in glass bottom petri dishes. The nanoscale spatial fluctuations (Σ) was calculated for each nucleus and averaged over 3 experiments (.n=~80 cells/group). SA1 loss decreased nuclear Σ (p<0.01), demonstrating that SA1 is key in maintaining nanoscale higher-order chromatin leading to more homogeneity of nuclear structures.