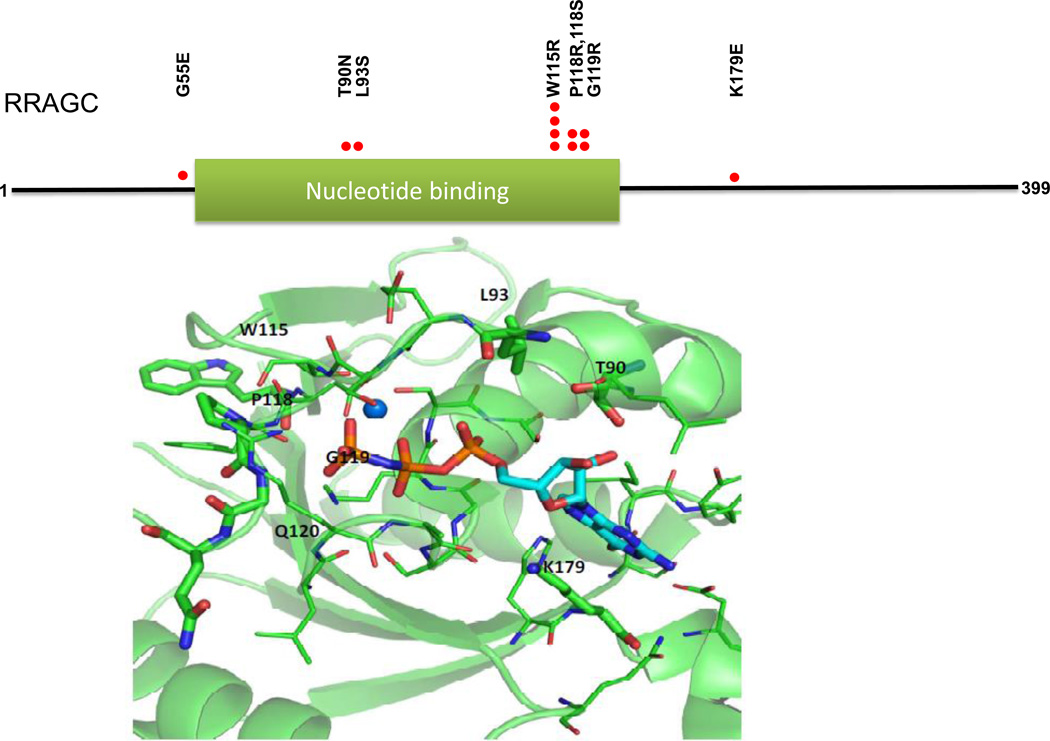
Figure 1.
FL-associated RRAGC mutations cluster on the protein surface surrounding the GTP/GDP binding site. Upper: Location of identified RRAGC missense mutations in a linear schema of RRAGC. Lower: Location of identified RRAGC missense mutations in the crystal complex (PDB:3LLU) of the RRAGC nucleotide binding domain (G-domain) bound with the nucleotide analog phosphoaminophosphonic acid guanylate ester (GNP), shown in stick representation. Amino acid residues undergoing mutation are labeled and shown in stick representation, while other residues within 4 angstroms of GNP are shown in line representation. A magnesium ion is shown as a sphere. The figure was generated with PyMOL.