Figure 2. Modeling of Fab2 steric hindrance to IN multimerization and DNA binding.
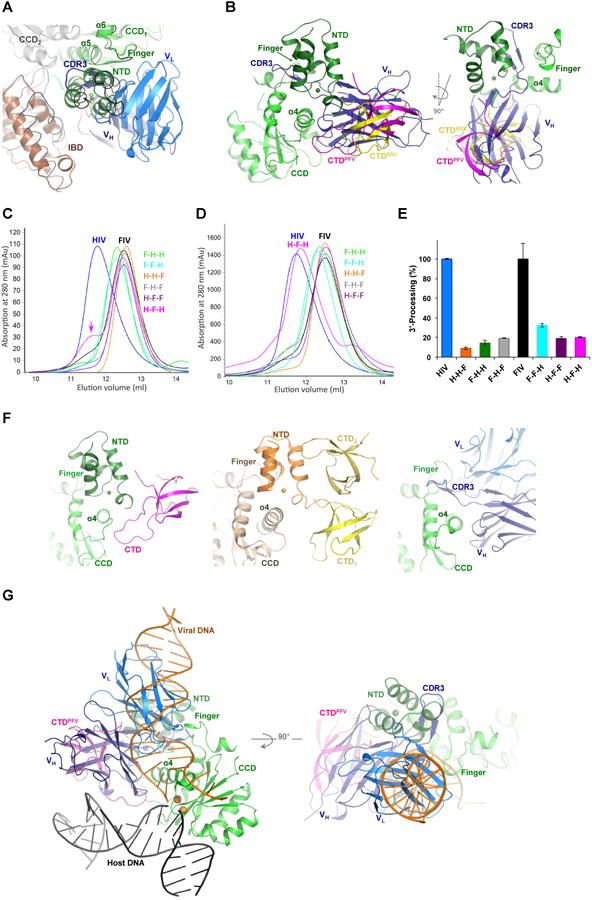
A) Superposition of HIV-1 IN-Fab2 complex to HIV-2 IN-IBD structure (3F9K). HIV-2 IN (gray and green cartoons) and IBD (brown cartoon) are shown together with Fab2 variable heavy (VH, deep blue) and light (VL, blue) chains. MVV IN-IBD complex structure (3HPH) is shown in black ribbon. B) Superposition of IN-Fab2 complex to PFV (3OS0) and RSV (5EJK) intasome structures showing PFV IN CCD (green), NTD (deep green), CTD (magenta) and the RSV CTD (yellow). C) Multimeric state of chimeric IN variants of HIV-1 “H” containing domains swapped from IN of FIV “F”, analyzed at low (0.25 mg/ml) and D) high (4.3 mg/ml) protein concentrations. E.g. H-H-F contains NTD and CCD of HIV-1 and CTD of FIV, while F-H-F contains NTD and CTD of FIV and CCD of HIV-1 and so forth. Color-code of IN variants in preserved through panel “E”. E) 3′-processing activity of IN variants. F) NTD and CTD wrapping of CCD a4 in intasome structures of PFV (3OS0, left) and RSV (5EJK, middle), which is mimicked by Fab2 binding (right panel). G) Superposition of IN-Fab2 complex to PFV strand-transfer complex structure (3OS0) showing PFV IN CCD (green), NTD (deep green) and CTD (magenta) bound to viral DNA (orange), host DNA (black) and two Mn2+ ions (gold spheres modeled from 3L2V).
