Abstract
SALL4 is aberrantly expressed in human myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). We have generated a SALL4 transgenic (SALL4B Tg) mouse model with pre-leukemic MDS-like symptoms that transform to AML over time. This makes our mouse model applicable for studying human MDS/AML diseases. Characterization of the leukemic initiation population in this model leads to the discovery that Fancl (Fanconi anemia, complementation group L) is down-regulated in SALL4B Tg leukemic and pre-leukemic cells. Similar to the reported Fanconi anemia (FA) mouse model, chromosomal instability with radial changes that can be detected in pre-leukemic SALL4B Tg bone marrow (BM) cells after DNA damage challenge. Results from additional studies using DNA damage repair reporter assays support a role of SALL4 in inhibiting the homologous recombination pathway. Intriguingly, unlike the FA mouse model, after DNA damage challenge, SALL4B Tg BM cells can survive and generate hematopoietic colonies. We further elucidated that the mechanism by which SALL4 promotes cell survival is through Bcl2 activation. Overall, our studies demonstrate for the first time that SALL4 has a negative impact in DNA damage repair, and support the model of dual functional properties of SALL4 in leukemogenesis through inhibiting DNA damage repair and promoting cell survival.
Keywords: SALL4, Fancl, Bcl2
Introduction
SALL4, a zinc-finger transcription factor of the SALL gene family, has been isolated based on its homology to a Drosophila region-specific homeotic gene, spalt. Human SALL4 mutations are associated with Duane-radial ray syndrome (DRRS, or Okihiro syndrome)(1–4). This condition is an autosomal-dominant disorder involving radial-sided hand anomalies in association with Duane syndrome (DS), which is a congenital disorder of eye movement characterized by strabismus. In addition, a novel SALL4 mutation has been identified in a large IVIC (Instituto Venezolano de Investigaciones Cientìficas) syndrome family by the Arias group(5). The IVIC syndrome is an autosomal-dominant condition with DRRS, leukocytosis and thrombocytopenia, which suggests that SALL4 may play a role in normal hematopoiesis(5). This observation has been further supported by additional studies(6). Knocking down SALL4 in normal human CD34+ hematopoietic stem/progenitors leads to impaired hematopoietic differentiation(6), and SALL4 plays a role in normal human hematopoiesis by affecting genes implicated in hematopoietic cell proliferation and differentiation such as HOXA9 and RUNX1(6).
In addition to its role in human development, SALL4 is an essential factor for embryonic stem (ES) cells. In recent years, multiple research groups(7–9), including ours(10), have demonstrated that SALL4 plays an essential role in the maintenance of ES cell pluripotency and self-renewal by interacting with other two key regulators in ES cells, Nanog and Oct4. Loss of SALL4 expression in ES cells results in down regulation of ES cell makers, such as Oct4, and spontaneous ES cell differentiation.
While SALL4 expression is down regulated during development, and not detectable in most human adult tissues, it is aberrantly expressed in high-grade MDS patients (11) and most AML patients(12). Loss-of-functional studies have demonstrated that SALL4 is essential for leukemic survival (13, 14), and SALL4 is being investigated as a new target in treating human MDS and AML. Direct evidence of the causative role of SALL4 in leukemogenesis has been demonstrated by studies from our laboratory in SALL4 transgenic (SALL4B Tg) mice, which developed pre-leukemic MDS-like features before subsequent AML transformation (12), making this murine model relevant to study human MDS/AML progression. Therefore, we aim to use this Tg model to study the mechanisms by which SALL4 promotes the progression from MDS to AML.
In this study, through gene expression profiling, we found Fancl (Fanconi anemia, complementation group L) to be down-regulated in leukemic initiating population as well as in the pre-leukemic bone marrow cells in the SALL4B Tg mice. Fancl is a component of the FA (Fanconi anemia) protein complex. Defects of the FA protein complex can lead to defective DNA damage repair. We tested the DNA damage repair capacity of SALL4B Tg bone marrow cells after challenge with the DNA damaging agent Mitomycin C (MMC) as well as by the use of homologous recombination (HR) and nonhomologous DNA end joining (NHEJ) reporter assays. Our results suggest that SALL4 can contribute to defective DNA damage repair through inhibition of the HR pathway. In addition, we found that Bcl2 (B-cell lymphoma 2) was up-regulated in the SALL4B Tg pre-leukemic bone marrow cells, which contributes to enhanced survival of these cells. Overall, in this report, we have explored the role of SALL4 in DNA damage repair and cell survival after DNA damage challenge and our findings provide new insight into SALL4-mediated leukemogenesis.
Results
Fancl was down-regulated in SALL4B transgenic (Tg) leukemic and pre-leukemic cells
In normal human bone marrow samples, SALL4B is expressed preferentially in the CD34+CD38− HSCs, and is down-regulated significantly in the CD34+CD38+ HPCs(6). By contrast, in leukemic samples, SALL4B is expressed at a higher level in the CD34+CD38+ HPCs than that in the CD34+CD38− HSCs (15). We previously reported the generation of SALL4B Tg mouse model (12). In this study, we further evaluated the pattern of SALL4B expression in these mice. We observed that SALL4B was expressed at a higher level in hematopoietic progenitor cell (HPC) populations than in phenotypically defined Lin-Sca-1+c-kit+ (LSKs, Supplementary Figure 1) enriched for hematopoietic stem cells (HSCs). This aberrant expression pattern of SALL4 is similar to what we have observed in human leukemia (15).
Based on previous data from these transgenic mice(12), we defined the pre-leukemic phase from 2 months until evolution of frank leukemia defined as leukemic phase, which usually occurred by 8 months. In order to determine whether the MDS-like cellular defect contributing to the leukemic phenotype originated at the stem or progenitor cell compartment, we analyzed the LSK and HPC sub-populations and correlated the numbers with the disease progression in SALL4B Tg mice. The total number of bone marrow cells was similar among wild type (WT), SALL4B Tg pre-leukemic, and leukemic animals. The percentages of both LSK and HPC populations were elevated significantly for pre-leukemic and leukemic stages in the SALL4B Tg mice compared with WT littermates (Supplementary Figure 2). The LSK and GMP cell populations were further expanded as the disease proceeded from the pre-leukemic to leukemic stages (Supplementary Figure 2). To identify which population can initiate the leukemic phenotype in a transplant model, we sorted the LSK and HPC subpopulations from primary leukemic SALL4B Tg donor mice and performed serial leukemic transplantations using a NOD-SCID mouse model as previously reported(12). As shown in Table 1, both LSKs and GMP cells gave rise to the leukemic phenotype in the recipients, indicating that both populations contain leukemia initiation activity.
Table 1.
Information of SALL4B-induced leukemic transplantations
| SALL4B Leukemic Donor Cells | Number of Donors | Number of Cells(1st transplant) | Number of Recipients (1st transplant) | Number of Cells(2nd transplant) | Number of Recipients (2nd transplant) | Observation Time (days) | AML Frequency |
|---|---|---|---|---|---|---|---|
| Whole Bone Marrow | 5 | 106 | 5 | 106 | 5 | 30–90 | 10/10 |
| Whole Spleen | 5 | 106 | 10 | 106 | 10 | 21–90 | 20/20 |
| LSK | 5 | 3.9×103 | 3 | 1.3×103 | 3 | 18–133 | 6/6 |
| GMP | 9 | 8×104 | 4 | 2.3×103 | 4 | 7–158 | 8/8 |
| CMP | 4 | 5–8×103 | 5 | _ | _ | Up to 240 | 0/5* |
| MEP | 4 | 1.5–8×104 | 5 | _ | _ | Up to 240 | 0/5* |
During the primary transplants, there were two cases of CMP and MEP donor cells that caused leukemic phenotypes in the recipient mice. However, FACS analysis was done on the leukemic recipients before the secondary transplant and showed only GMP population present, which suggested that these CMP and MEP cases were contaminated with GMPs.
To investigate the molecular mechanism of SALL4’s role in the development of leukemia, and to characterize the genetic changes that occur during the transition from a normal hematopoietic cells to leukemia-initiating cells (LIC), we isolated the Leukemia (L)-GMPs and L-LSKs from three independently transplanted NOD-SCID recipients, then compared their gene expression profiles to those populations isolated from pre-leukemic SALL4B transgenic mice, control NOD-SCID (NOD) and wild type (WT) littermates. Unsupervised analysis demonstrated that the global gene expression profiles of SALL4-GMPs (L-GMP and P-GMP) or LSKs (L-LSKs and P-LSK) were similar to those from the respective controls (NOD-GMP and WT-GMP or NOD-LSK and WT-LSK) (Figure 1a).
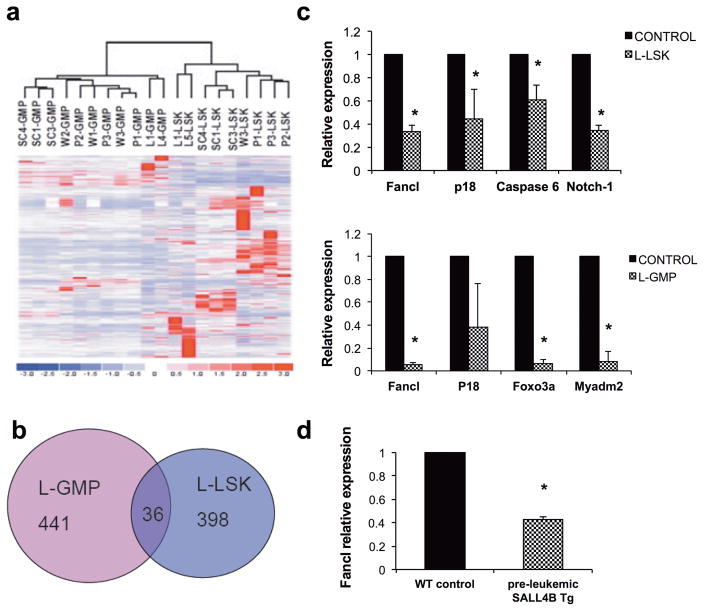
Figure 1. Fancl was down-regulated in SALL4B Tg leukemic and pre-leukemic bone marrow cells.
(A) Hierarchical clustering using the 9,100 filtered probe sets demonstrated that SALL4B pre-leukemic GMP (P1-3 GMP) and leukemic GMP (L1-GMP and L4-GMP) shared a gene expression profile most similar to WT control GMP (W1-3 GMP were from BL6 mice, and SC1, 3 and 4 GMP were from NOD-SCID mice). Similarly, the SALL4B pre-leukemic LSK and leukemic LSK shared a similar gene expression profile to WT control LSK. (B) Venn diagram showing the overlap gene probes between SALL4B L-GMP and SALL4B L-LSK. Only 36 genes were common to the two LIC populations. (C) Validation of SALL4B specific target genes. qRT-PCR analysis of gene expression in SALL4B L-GMP and SALL4B L-LSK when compared to control normal GMP and LSK respectively were performed. Measurements were from individual transplanted recipients, each performed in duplicate. Error bars indicate SD. (D) qRT-PCR analysis showed Fancl was down-regulated in pre-leukemic SALL4B transgenic mice whole bone marrow cells when compared to those from WT control. Each group has 5 mice, Error bars indicate SD. * indicate p<0.05.
Further analysis using dChip revealed an expression signature for the L-LSK, which consists of both up-regulated (93 probe sets) and down-regulated genes (341 probe sets). Similarly an expression signature for the L-GMP consisted of up-regulated (83 probe sets) and down-regulated genes (394 probe sets). There were 36 dysregulated genes shared between these two LIC populations (Figure 1b and supplementary Table 2). Among these, the altered expression of several potentially relevant genes was verified by qRT-PCR. In the SALL4B L-LSK population, Fancl (p<0.001), p18 (p=0.044), Caspase 6 (p=0.015), and Notch1 (p<0.001) were all down-regulated when compared to those from control WT LSKs. In L-GMPs, Fancl (p<0.001), Foxo3a (p<0.001) and Myadm2 (p<0.001) were confirmed to be down-regulated when compared to those from control WT GMPs. Thus, Fancl was down-regulated in both LIC populations (Figure 1c).
FANCL belongs to the FA (Fanconi anemia) protein complex. FA is an inherited bone marrow failure syndromes resulting from deficiency or mutation of one of 19 proteins (FA complementation groups A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P, Q, R, S and T, http://www.rockefeller.edu/fanconi/) that are involved for DNA repair. As a result, 20% or more of FA patients develop cancer, most often AML, and 90% of those develop bone marrow failure and/or some features of MDS by the age of 40 (16). Thus, we hypothesized that the pre-leukemic and MDS phenotypes of SALL4B Tg mice was linked to reduced Fancl expression. To test this hypothesis, we examined the expression of Fancl by quantitative real-time RT-PCR in five pairs of bone marrow cells from age-matched WT and SALL4B pre-leukemic Tg mice. Fancl expression was significantly down-regulated in SALL4B pre-leukemic Tg mice when compared to that of WT (Figure 1d).
SALL4B Tg mice had HR deficiency but not NHEJ in DNA damage repair
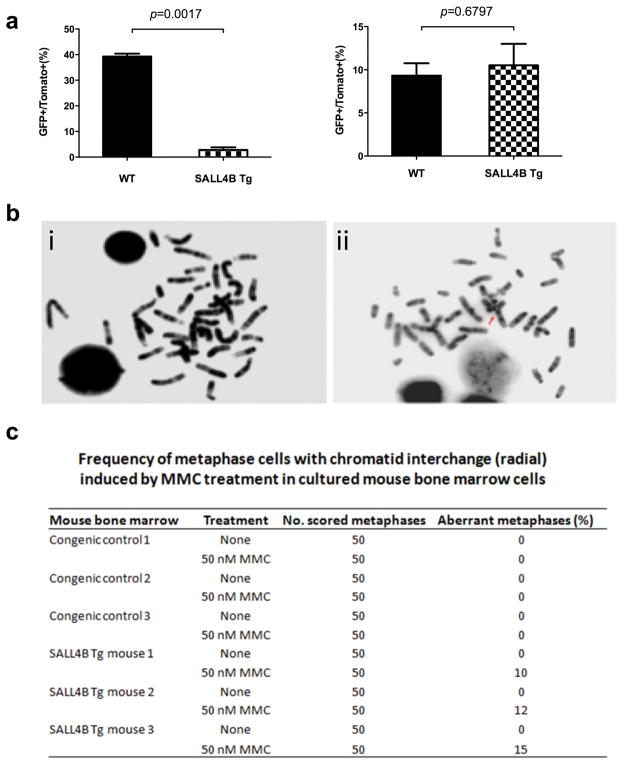
FANCL is an E3 Ubiquitin ligase protein that mediates the monoubiquitination of FANCD2, a key step in activating the downstream FA pathway that triggers the subsequent DNA damage repair process. To determine if SALL4 plays a role in DNA damage repair through its role in regulating Fancl, we utilized HR and NHEJ reporter systems(17–21) to explore the effect(s) of SALL4 expression on specific DNA damage repair pathway(s). We first created DNA double-strand breaks (DSB) in cells transfected with HR or NHEJ reporter cassette in vitro by I-SceI restriction digest. As shown in Figure 2a, the HR repair efficiency was significantly decreased in the pre-leukemic SALL4B Tg lineage negative cells when compared with WT control (p=0.0017). There was no difference for NHEJ repair efficiency (p=0.6797). To confirm the direct role of SALL4B in HR, we over-expressed SALL4B in a human leukemia cell line KBM5, (Supplementary Figure 3a) and observed that the HR repair was significantly decreased in these cells (p=0.0034) compared to the WT cells (Supplementary Figure 3b) while there was no difference in NHEJ repair (p=0.3401) (Supplementary Figure 3c). More importantly, overexpressing FANCL could rescue the HR repair defect mediated by ectopic SALL4 expression (Supplementary Figure 4). In addition, overexpressing SALL4B in KBM5 and H1299 cells led to decreased monoubiquitination of FANCD2 (Supplementary Figure 5).
Figure 2. SALL4B transgenic mice had HR deficiency but not NHEJ in DNA damage repair, and SALL4B transgenic mice had chromosomal instability after MMC treatment.
(a) HR and NHEJ reporter assay was performed on pre-leukemic SALL4B Tg mice and WT control. (b) & (c) SALL4B transgenic mice bone marrow cells were more sensitive to DNA damage drug MMC treatment by producing more chromosomal aberrations (radial changes) (ii) than WT control (i).
SALL4B Tg mice displayed chromosomal instability after MMC treatment
Cells defective in the FA pathway display an increase frequency of chromosomal breaks after challenged with the DNA crossl-linking drug MMC(22) and chromosome aberrations such as chromatid interchange triradial and quadriradial are typically observed in MMC-treated cells from FA patients(23) or FA mice(24). Since the SALL4B Tg mice showed decreased Fancl expression level, we next examined whether SALL4B Tg pre-leukemic mice had an unstable chromosomal phenotype after MMC challenge. SALL4B Tg mouse bone marrow cells displayed typical FA-like chromosomal abnormalities such as chromatid interchange radial changes at a rate of 10–15/per 50 counted cells (Figure 2b and Figure 2c) after MMC treatment, while cells from WT mice showed no visible chromosomal abnormalities. Intriguingly, older (>8 month) SALL4 Tg mice had statistically significantly more abnormal chromosomal abnormalities per cell than the younger SALL4 Tg mice (≤ 8 month) after MMC challenge (Supplementary Figure 6).
High SALL4 expression level was correlated with complex karyotype in MDS/AML patients
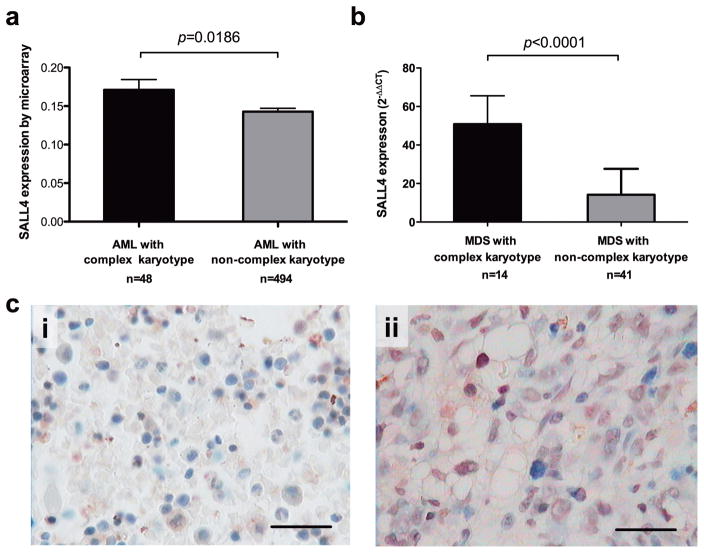
To examine whether SALL4 is also involved in chromosomal instability in human primary MDS/AML patients, we evaluate the correlation between SALL4 expression and chromosome karyotypes of these samples. Using publicly available datasets (GSE13159) (25, 26), we found that the expression of SALL4 in AML patients with complex karyotype (equal to or more than three aberrant karyotypes) was significantly higher (p=0.0186) than that in MDS patients with normal karyotype (Figure 3a). In addition, re-analysis of our prior collection of MDS patient samples(11) showed higher SALL4 RNA (Figure 3b) and protein (Figure 3c) expression in MDS patients was associated with the complex karyotype.
Figure 3. High SALL4 expression level was correlated with complex karyotype in primary MDS patient samples.
(a) In an AML cohort of 542 patients (GSE13159) analyzed by microarray, the expression of SALL4 in AML patients with complex karyotype was significantly higher (p=0.0186) than that in MDS patients with normal karyotype. (b) In a cohort of 55 MDS patients analyzed by qRT-PCR, SALL4 in MDS patients with complex karyotype was significantly higher (p<0.001) than that in MDS patients with normal karyotype. (c) Representative SALL4 protein expression in MDS patient with complex karyotype (ii) was higher than that in MDS patient with normal karyotype (i), analyzed by IHC.
Since TP53 mutation/alternation is the most common biomarker associated with complex karyotype in human MDS patients (27), we next examined whether there was an association between SALL4 expression and TP53 mutation. Since TP53 mutation in cancer cells leads to prolonged half-life of the protein, IHC has been used to detect mutant TP53(28). Twenty MDS cases were selected for the evaluation of SALL4 expression and TP53 mutation. Overall, sixteen out of twenty cases (80%) showed consistency for TP53 and SALL4 based on the positive or negative IHC results (Table 2 and Supplementary Figure 7). SALL4 expression was positively correlated with TP53 expression/mutation in these MDS patients (contingency coefficient r=0.52, p<0.05). Future study with a larger cohort of MDS patients is required to validate this result.
Table 2.
IHC results for TP53 and SALL4 in 20 MDS patients
| No | Karyotype | TP53 | SALL4 |
|---|---|---|---|
| 1 | -5q22,q35 | - | - |
| 2 | Normal | - | - |
| 3 | Complex | 2+ | 3+ |
| 4 | Complex | 3+ | 1+ |
| 5 | Complex | 3+ | 1+ |
| 6 | Normal | - | - |
| 7 | Complex | - | - |
| 8 | +8 | 1+ | 4+ |
| 9 | Complex | - | 2+ |
| 10 | +19p13 | 1+ | - |
| 11 | +19p13 | 1+ | 1+ |
| 12 | Normal | - | - |
| 13 | Normal | - | 4+ |
| 14 | Complex | 2+ | 3+ |
| 15 | Complex | 3+ | 3+ |
| 16 | Normal | - | - |
| 17 | Normal | - | - |
| 18 | Normal | - | 3+ |
| 19 | ?-8q23,?22q11 | - | - |
| 20 | +21 | - | - |
TP53 was positive in 71.4% (5/7) MDS patients with complex karyotype and 23.1% (3/13) in non-complex karyotype. SALL4 was positive in 85.7% (6/7) MDS patients with complex karyotype and 30.8% (4/13) in non-complex karyotype.
Consistency in terms of positive or negative for TP53 and SALL4 is 80%(16/20).
SALL4B Tg mice bone marrow cells had increased Bcl2 expression
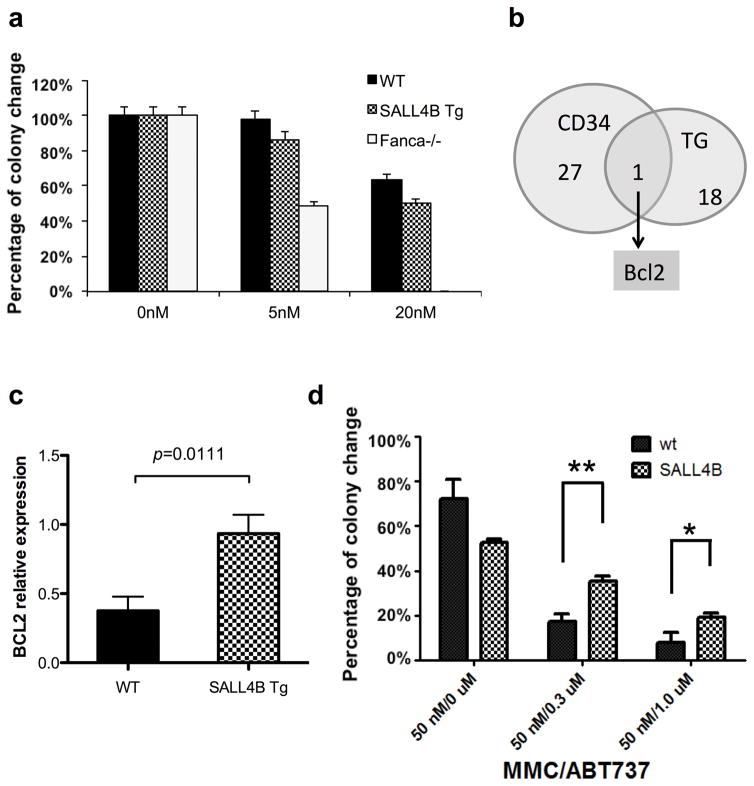
Similar to other mouse models of FA, Fanca−/− mice demonstrate hypersensitivity to bone marrow damage with increased stress-induced apoptosis and reduced marrow progenitor cells after MMC treatment(29). To evaluate whether SALL4B Tg mice showed a similar phenotype, we performed colony forming unit (CFU) assays after MMC treatment of both SALL4B Tg and Fanca−/− BM cells. Surprisingly, despite our observation of abnormal chromosomal changes after MMC treatment in our SALL4B Tg pre-leukemic BM cells (Figure 2c), unlike the Fanca−/− mice, we observed no increased sensitivity to MMC, with CFU colonies in SALL4B Tg BM cells even at concentrations as high as 20 nM (Figure 4a). One explanation for this difference in response to MMC is the possibility that SALL4B cells express increased anti-apoptosis signals. To explore this hypothesis, we next performed gene expression profiling on SALL4B Tg pre-leukemic BM cells. Overall, 67 genes were up-regulated, and 55 genes were down-regulated in SALL4B Tg pre-leukemic BM cells when compared to the WT controls (Supplementary Table 3). A comparison with the gene expression profiles of SALL4B Tg pre-leukemic BM cells with our previous studies on human CD34+ cells(6) revealed BCL2 was the overlap gene in both data sets (Figure 4b and supplementary table 4), suggesting that BCL2 is a conserved target of SALL4 in mice and human. We further confirmed the microarray data by qRT-PCR assay and showed that Bcl2 expression was significantly increased in SALL4B Tg mouse BM cells compared with WT controls (p=0.0111) (Figure 4c). Similarly, overexpressing SALL4 in KBM5 cells led to increased BCL2 expression with decreased FANCL expression (Supplementary Figure 8).
Figure 4. SALL4B Tg mice bone marrow cells had better survival ability than FA mice by up-regulating Bcl2.
(a) Colony forming assay on SALL4B Tg mice bone marrow cells after MMC treatment. WT mice and Fanca−/− mice were used as negative and positive control respectively. For Fanca−/− mice, colony number decreased significantly after 5 nM MMC treatment and no colony was observed after 20 nM MMC treatment. Unlike the Fanca−/− model, SALL4B Tg mice bone marrow cells can still survive and form colonies after 20 nM MMC treatment. (b) Venn diagram showing the overlap apoptosis-related gene probes between SALL4B pre-leukemic bone marrow cells and human CD34+ cells. BCL2 was the only common gene. (c) qRT-PCR result confirmed that Bcl2 was significantly increased (p=0.0111) in SALL4B Tg mouse bone marrow than WT control. (d) SALL4B Tg bone marrow cells were more resistant to BCL2 inhibitor ABT-737 treatment after DNA damage drug MMC challenge. Colony formation assay showed that SALL4B BM cells were less sensitive to the treatments of Bcl-2 inhibitor ABT-737 than the WT BM cells. Cells were seeded in methylcellulose medium containing cytokines (M3434) containing 50 nM of MMC in the absence or presence of o.3 μM or 1 μM of ABT-737. Colonies were scored after 7 days. Data are the mean±SD of colony numbers expressed as a percentage of DMSO-treatment (0 μM of MMC). Error bars represent SD of WT BM (n=3) and SALL4B BM (n=3). *P<0.05; **P<0.01
To further confirm that changes in Bcl2 mRNA results in functional alterations, we treated mouse BM cells with a BCL2 inhibitor ABT-737 and MMC and performed CFU assays. The number of colonies from SALL4B Tg BM cells treated with both MMC and ABT-737 were significantly higher than that from WT controls treated similarly (Figure 4d and Supplementary Figure 9). Together, these results suggest that SALL4 promotes cell survival in part through the BCL2 pathway.
Discussion
Human MDS is a group of clonal HSC disorders characterized by cellular dysplasia and refractory cytopenias as a result of ineffective hematopoiesis. MDS is mostly seen in the elderly, with over 10,000 new cases per year in the US alone. As the population ages, the role of this disease in limiting life expectancy will become more significant. About 30 to 40 percent of MDS patients progress to AML with a poor prognosis and short life expectancy. However, it remains unclear what mechanism(s) drives the progression of MDS to AML, the understanding of which will enable development of novel strategies in treating these patients. Few murine models have been generated that mimic human MDS/AML disease progression. We have previously reported that SALL4 is aberrantly expressed in human MDS/AML (11, 12), and SALL4B Tg mice develop MDS-like features before progressing to AML (12). Therefore, we propose to use the SALL4B Tg murine model to search for novel insight(s) in this human disease.
Here, we first explored the role of SALL4 in leukemogenesis by monitoring the changes of gene expression profiles from the hematopoietic sub-populations and LIC populations during disease progression in the SALL4B Tg mouse model. Thirty-six genes are shared by these two LIC populations, one of which is Fancl, a gene that encodes an E3 ubiquitin ligase in the FA protein complex which plays a crucial role in DNA repair and genomic stability maintenance (30, 31). Interestingly, deregulation of FANCL has been reported in AML patients (32, 33). We demonstrated that the expression of Fancl is significantly down-regulated in pre-leukemic SALL4B Tg mouse BM cells. SALL4 probably affects the expression of FANCL gene indirectly since no binding peaks were observed at the promoter region of this gene in our previous global ChIP-Chip studies (6, 14).Several FA mouse models are available that share a phenotype of mild ineffective hematopoiesis and hypersensitive to the DNA cross-linking agent MMC (22, 34, 35). In the SALL4B Tg mouse model, we have observed abnormal chromosomal changes after MMC treatment that are similar to those seen in FA mice that correlated with decreased Fancl expression. We also demonstrate using reporter assays that HR DNA damage repair pathway is deficient in SALL4B Tg mouse BM cells. Similar results that correlate SALL4 expression with chromosomal abnormalities were also observed in human samples as primary MDS and AML patients with complex karyotype have higher SALL4 expression level. Finally, we demonstrated that over expression of SALL4 in KBM5 cell line led to impaired HR repair ability, which can be rescued by overexpressing FANCL.
Intriguingly, unlike the Fanca−/− mice and other murine models of FA, SALL4B Tg mouse BM cells did not show hypersensitivity to progenitor cell growth with MMC treatment. Our previous study demonstrated that SALL4 plays a key role in leukemic cell survival by affecting multiple pathways, including the direct activation of anti-apoptotic gene Bcl2(14). Our current study demonstrated that Bcl2 is up-regulated in SALL4B Tg pre-leukemic mice, which may account for the enhanced survival of BM progenitors of SALL4B Tg cells after MMC treatment since these cells are more resistant to the BCL2 inhibitor ABT-737 treatment.
In summary, our studies indicate that SALL4 can negatively affect the DNA damage repair processes, at least in part, through repressing Fancl. In addition, SALL4 can promote cell survival by activating the anti-apoptosis gene Bcl2. Taken together; it is possible that the SALL4B Tg mice are more prone to accumulate secondary mutations, which may lead to their progression from MDS into the AML stage. Overall, our studies suggest that SALL4 has dual functional roles in inhibiting DNA damage repair and promoting cell survival following genotoxic stress. Our findings provide novel insight on the pathogenesis of SALL4 in MDS/AML disease progression.
Materials and Methods
Mice and serial transplantation
SALL4B Tg mice were generated as previously described (12, 36). Both male and female mice, age from 3 month to 12 months were used. Serial transplantation was also previously described (36). Briefly, whole bone marrow, spleen, sorted LSKs, (Lin-Sca-1+c-kit+), common myeloid progenitors (CMPs, Lin-Sca-1-c-kit+CD34+ FcγRII/IIIlow), granulocyte-macrophage progenitors (GMPs, Lin-Sca-1-c-kit+CD34+ FcγRII/III+), and megakaryocyte erythroid progenitors (MEPs, Lin-Sca-1-c-kit+CD34− FcγRII/III−) from SALL4-induced leukemic mice or primary recipient mice were transplanted into NOD/SCID mice by tail vein injection. The transplantation cell-dose ranges were shown in Table 1. The Fanca knockout mice were described previously (29). All animal work has been conducted in accordance to relevant national and international guidelines, as well as the recommendations of the Weatherall report, and the mice are housed at Children’s Hospital Boston. The sample size estimate is based on a 80% predication power, with a 0.05 significance level to detect a 1.00 standard deviation (SD) difference between the two study groups. Randomization or blinding is not applicable to our studies.
Fluorescence-activated cell-sorting analysis (FACS)
Hematopoietic stem and progenitor cells were isolated from mouse bone marrow cells as described previously (37). Briefly, mouse bone marrow cells were stained for the following antibodies: Lineage marker (CD3, CD4, CD8, Gr-1, CD19, B220 and Ter119) (Invitrogen, Caltag Laboratories), Sca-1, c-kit, CD34 and FcγR (Fc gamma receptors) II/III (BD Biosciences PharMingen, San Diego, CA). After 30 minutes staining at 4°C, cells were washed 3 times with PBS and resuspended in 1×PBS with 2% heat-inactivated FBS and 1μg/ml PI. Then the cells were analyzed or sorted using MoFlo cell sorter (Dako, Fort Collins, Colorado).
Polymerase chain reaction (PCR) and quantitative reverse transcription-PCR (qRT-PCR)
PCR and qRT-PCR were performed as previously described (12). Briefly, a PCR kit (Qiagen, Valencia, CA) was used to genotype the SALL4B Tg founder mice and transmission of the transgene. Genomic DNA was purified from a mouse tail using a high-quality DNA kit (Gentra Systems, Minneapolis, MN). The human SALL4B primer sequences were showed in supplementary table 1. Total RNA was isolated using a phenol-free and filter-based RNA isolation system (Qiagen) digested with DNase I to remove DNA contamination. Primer sequences for qRT-PCR were designed using Primer Express® software (Applied Biosystems, Foster City, CA) and were listed in supplementary table 1. All reactions were performed in an ABI-7000 sequence detection system using TaqMan PCR core reagents according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA). PCR amplification was carried out in a 50 μl final volume containing 1×PCR buffer, 1 μM of each primer, and 10 μl of RNA at 95°C for 10 min followed by 45 cycles at 95°C for 15 sec and then at 60°C for 1 min. For each sample, expression of the GAPDH gene was used to normalize the amount of investigated transcript.
Gene expression profiling
Total RNA of various populations or whole bone marrow cells (2–5 x104 cells/population) from wild type (WT), NOD-SCID and the NOD-SCID mice with leukemic cell-transplants was extracted using Trizol reagent (Invitrogen Corp, Carlsbad, CA, USA). The amount of extracted RNA was quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Welmongton, DE, USA). RNAs (50 ng) were reverse- transcribed to cDNAs, then amplified by single primer isothermal amplification (SPIA). Finally, the amplified products (3.75 μg) were fragmented and biotin labeled by OvationTM RNA Amplification System V2 and FL-OvationTM cDNA Biotin Module V2 kits (NuGEN Technologies, San Carios CA, USA) according to the manufacturer’s instructions. Labeled cDNAs were hybridized to an Affymetrix Mouse Genome 430A 2.0 Array and detected by the Microarray Core Facility at Dana-Farber Cancer Institute. The gene expression data were deposited in the European Bioinformatics Institute ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) with accession number E-MEXP-2072.
Data analysis
Data were analyzed by dChip (http://biosun1.harvard.edu/complab/dchip/) and GSEA 2.0 algorithm (http://www.broad.mit.edu/gsea/). With dChip the raw expression data was normalized to account for differences in chip intensities and then filtered using 0.4<standard deviation/mean<1000, presence call% in the arrays used ≥ 20% and the expression level ≥ 20 in ≥ 20% samples. Hierarchical clustering was performed among the filtered genes. For comparisons of gene expression between two groups, the transcripts were considered as up- or down-regulated when their transcript levels in SALL4B Tg mice, as compared to multiple control animals, were increased or decreased more than 1.5 fold, p-value<0.05, and lower confidence bound at >90%.
Mitomycin C (MMC) clonogenic assays
The MMC cologenic assays have been described previously for FA mice (22). Briefly, bone marrow cells were harvested from age-matched wild type (WT) or SALL4B Tg mice. Low-density bone marrow mononuclear cells (BMMCs) were isolated by density gradient centrifugation using Histopaque 1.083 g/cm3 (Sigma, St Louis, MO). For clonogenic assays, 10,000 mononuclear cells were plated in triplicates in methylcellulose (Methocult; Stem Cell Technologies, Vancouver, Canada) supplemented with 30% fetal calf serum, 2 mmol/l L-glutamine (Hyclone, Logan, UT), 200 U/ml penicillin/streptomycin (Hyclone), 100 mmol/l β-mercaptoethanol (Thermo Fisher, Waltham, MA), 1% bovine serum albumin (Roche, Indianapolis, IN), and 100 ng/ml recombinant rat stem-cell factor, 100 ng/ml murine IL3, 4 U/ml Epogen (Amgen). 5 or 20μM of MMC (Bedford Laboratories Bedford, OH) were added. The plates were incubated for 7 days in a humidified atmosphere (37 °C, 5% CO2), and colonies were counted on day 7.
Similarly, whole bone marrow cells were harvested from age-matched WT or SALL4B Tg mice for MMC and/or ABT737 colony forming assay. 10,000 mononuclear cells were plated out in triplicate in methylcellulose (MethoCult™ GF M3434; Stem Cell Technologies, Vancouver, Canada). The cells were treated with 50 nM of MMC alone (Bedford Laboratories Bedford, OH), 0.3 μM or 1.0 μM of ABT-737 alone (provided by Abbott Laboratories)(38), or MMC and ABT-737 together. The plates were incubated as above and colonies were counted on day 7.
Chromosomal spread
Bone marrow cells were cultured the same way as described above for the MMC colony formation assay. At day 5, cultured bone marrow cells were exposed to 50 nM of MMC and cultured for another 24 hours. Before harvest, Colcemid was added to a final concentration of 0.1 ug/ml in for 45 min. Chromosome spreading were prepared for metaphase analysis as previously described(39).
Homologoous recombination (HR) and nonhomologous DNA end joining (NHEJ) reporter assay
Lineage negative SALL4B Tg and WT mouse bone marrow cells and leukemia cell line KBM5 cells (a gift from Dr. Ma’s Lab, and used in our previous report(15)) were used to perform HR and NHEJ reporter assays (17–21). Plasmids containing HR or NHEJ reporter cassettes were linearized by I-SceI restriction enzyme (R0694L, NEB) and purified using QIAquick PCR Purification Kit (28106; Qiagen). Lineage negative bone marrow cells or KBM5 cells were electroporated with 0.5 μg of the NHEJ reporter construct or 2 μg of the HR reporter constructs, along with 0.1 μg of the tdTomato expression vector to normalize for differences in transfection efficiency. Electroporations were performed using the Amaxa Nucleofector (Walkersville, MD). After transfection, lineage negative cells were cultured in Serum-Free Expansion Medium (09600, STEMCELL) containing cytokines (10 ng/ml IL-3, 10 ng/ml IL-6, 50 ng/ml TPO and 50 ng/ml SCF). KBM5 cells were cultured in RPMI 1640 with 20% FBS. GFP gene was normally inactive in the transfected HR or NHEJ constructs. Only when HR or NHEJ events have occurred, can an active GFP gene be reconstituted. Expression of GFP and Tomato was analyzed by FACS (BD FACSCalibur) 72h after transfection. The ratio of GFP+ to Tomato+ cells was used as a measure for repair efficiency.
Western blots of FANCD2
Whole-cell extracts were immunoblotted with anti-FANCD2 antibody (NB100-182, NOVUS, USA). The upper band (slightly larger form) is monoubiquitinated FANCD2 (FANCD2-L), and the lower band is unubiquitinated FANCD2 (FANCD2-S) (40, 41). The ratio of FANCD2-L to FANCD2-S (L/S) was calculated by densitometry.
Primary human MDS/AML
The gene expression data on a cohort of 542 AML patients were downloaded from publicly available Gene Expression Omnibus (GEO) dataset under series accession number GSE13159 (25, 26). In addition, for our studies, we used primary human MDS/AML samples, which was described before (11). Briefly, bone marrow samples from 55 newly diagnosed MDS patients were collected in Beijing from December 2009 to December 2011. Another cohort of 20 MDS patient bone marrow tissues was collected in Peking Union Medical College Hospital from 2013 to 2015 and used for analysis of SALL4 and TP53 protein expression. This study was approved by the institutional review board of Peking Union Medical College Hospital and informed consent was obtained from all subjects.
Immunohistochemistry (IHC) staining of human MDS samples
IHC staining was performed on human MDS samples according to standard techniques as previously described (12, 42). Briefly, paraffin-embedded sections were first de-paraffinized and hydrated. Then the slides were microwaved in 10 mM sodium citrate for 15 minutes to retrieve the antigen. After cooling, the slides were blocked in PBST (PBS with 0.3% Triton) with 2% dry milk for 30 minutes. Tissue slides were then incubated with SALL4 antibody (ab29119, Abcam) or p53 (05-224, clone BP53−12, Millipore) overnight at room temperature. After 3 washes in PBST, slides were incubated with the secondary antibody (ZB-2301, ZSGB-BIO) for 1 hour at room temperature. After 2 more washes, the slides were incubated in ABC reagent (Vectastain) and developed in DAB solution. Hematoxylin (Fisher Scientific) was used to counterstain the slides. Cells were considered to be positive for SALL4 when they showed definitive nuclear staining.
Statistical analysis
Statistical analyses were performed using Graphpad Prism 5. P-values of <0.05 were considered statistically significant. Data were presented as means ± standard deviation. The variance between the groups that is in general similar being statistically compared. Unpaired t-test or Mann–Whitney test was used to study difference between two groups. Chi-Square Test was used to analyze the correlation between SALL4 and TP53 expression.
Supplementary Material
Acknowledgments
L Chai and W Cui designed the research, analyzed data and wrote the paper. F Wang and C Gao performed the research experiments, analyzed data and wrote the paper. J Lu and H Tatetsu performed the research experiments and analyzed the data. D Williams provided Fanca−/− mice and edited the paper. L Muller performed Fanca−/− mice related experiments. We also want to thank Dr. S Xiao for providing DNA repair reporter plasmids, X Yao, and D Neuberg for statistic analysis, X Tian in assisting animal work, and N Kong in editing of the manuscript. This work was supported in part through NIH R03CA184531 and PO1HL095489, funding from Lymphoma and Leukemia Society (No. SLP-8004-14 and TRP-6482-16) and V cancer research Foundation (to L Chai), by the National Natural Science Foundation of China (NSFC) - 81071418 & 81472029 (to W Cui), and China Scholarship Council (No.201206210154) (to F Wang).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Al-Baradie R, Yamada K, St Hilaire C, Chan WM, Andrews C, McIntosh N, et al. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. American journal of human genetics. 2002 Nov;71(5):1195–9. doi: 10.1086/343821. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borozdin W, Wright MJ, Hennekam RC, Hannibal MC, Crow YJ, Neumann TE, et al. Novel mutations in the gene SALL4 provide further evidence for acro-renal-ocular and Okihiro syndromes being allelic entities, and extend the phenotypic spectrum. J Med Genet. 2004 Aug;41(8):e102. doi: 10.1136/jmg.2004.019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohlhase J, Schubert L, Liebers M, Rauch A, Becker K, Mohammed SN, et al. Mutations at the SALL4 locus on chromosome 20 result in a range of clinically overlapping phenotypes, including Okihiro syndrome, Holt-Oram syndrome, acro-renal-ocular syndrome, and patients previously reported to represent thalidomide embryopathy. J Med Genet. 2003 Jul;40(7):473–8. doi: 10.1136/jmg.40.7.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohlhase J, Heinrich M, Schubert L, Liebers M, Kispert A, Laccone F, et al. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet. 2002 Nov 1;11(23):2979–87. doi: 10.1093/hmg/11.23.2979. [DOI] [PubMed] [Google Scholar]
- 5.Paradisi I, Arias S. IVIC syndrome Is caused by a c.2607delA mutation in the SALL4 locus. Am J Med Genet A. 2007 Feb 15;143(4):326–32. doi: 10.1002/ajmg.a.31603. [DOI] [PubMed] [Google Scholar]
- 6.Gao C, Kong NR, Li A, Tatetu H, Ueno S, Yang Y, et al. SALL4 is a key transcription regulator in normal human hematopoiesis. Transfusion. 2013 May;53(5):1037–49. doi: 10.1111/j.1537-2995.2012.03888.x. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren M, Wang W, Spiden S, Chen-Murchie D, Tannahill D, Steel KP, et al. A Sall4 mutant mouse model useful for studying the role of Sall4 in early embryonic development and organogenesis. Genesis. 2007 Jan;45(1):51–8. doi: 10.1002/dvg.20264. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, et al. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem. 2006 Aug 25;281(34):24090–4. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006 Nov 16;444(7117):364–8. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nature cell biology. 2006 Oct;8(10):1114–23. doi: 10.1038/ncb1481. eng. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Guo Y, Chen Q, Yang Z, Ning N, Zhang Y, et al. Stem cell factor SALL4, a potential prognostic marker for myelodysplastic syndromes. Journal of hematology & oncology. 2013;6(1):73. doi: 10.1186/1756-8722-6-73. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Y, Cui W, Yang J, Qu J, Di C, Amin HM, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006 Oct 15;108(8):2726–35. doi: 10.1182/blood-2006-02-001594. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Chai L, Liu F, Fink LM, Lin P, Silberstein LE, et al. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proceedings of the National Academy of Sciences of the United States of America. 2007 Jun 19;104(25):10494–9. doi: 10.1073/pnas.0704001104. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, et al. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008 Aug 1;112(3):805–13. doi: 10.1182/blood-2007-11-126326. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Ma Y, Kong N, Alipio Z, Gao C, Krause DS, et al. Dissecting the role of SALL4, a newly identified stem cell factor, in chronic myelogenous leukemia. Leukemia. 2011 Jul;25(7):1211–3. doi: 10.1038/leu.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med. 2010 May 20;362(20):1909–19. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seluanov A, Mao Z, Gorbunova V. Analysis of DNA double-strand break (DSB) repair in mammalian cells. Journal of visualized experiments : JoVE. 2010;(43) doi: 10.3791/2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia. 2009 Jul;11(7):683–91. doi: 10.1593/neo.09312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao Z, Seluanov A, Jiang Y, Gorbunova V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America. 2007 Aug 7;104(32):13068–73. doi: 10.1073/pnas.0702410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proceedings of the National Academy of Sciences of the United States of America. 2004 May 18;101(20):7624–9. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell cycle. 2008 Sep 15;7(18):2902–6. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller LU, Milsom MD, Kim MO, Schambach A, Schuesler T, Williams DA. Rapid lentiviral transduction preserves the engraftment potential of Fanca(−/−) hematopoietic stem cells. Molecular therapy : the journal of the American Society of Gene Therapy. 2008 Jun;16(6):1154–60. doi: 10.1038/mt.2008.67. [DOI] [PubMed] [Google Scholar]
- 23.Oostra AB, Nieuwint AW, Joenje H, de Winter JP. Diagnosis of fanconi anemia: chromosomal breakage analysis. Anemia. 2012;2012:238731. doi: 10.1155/2012/238731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Kuang Y, Montes De Oca R, Hays T, Moreau L, Lu N, et al. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood. 2001 Dec 1;98(12):3435–40. doi: 10.1182/blood.v98.12.3435. [DOI] [PubMed] [Google Scholar]
- 25.Kohlmann A, Kipps TJ, Rassenti LZ, Downing JR, Shurtleff SA, Mills KI, et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. British journal of haematology. 2008 Sep;142(5):802–7. doi: 10.1111/j.1365-2141.2008.07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010 May 20;28(15):2529–37. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bejar R, Stevenson KE, Caughey B, Lindsley RC, Mar BG, Stojanov P, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014 Sep 1;32(25):2691–8. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alsner J, Jensen V, Kyndi M, Offersen BV, Vu P, Borresen-Dale AL, et al. A comparison between p53 accumulation determined by immunohistochemistry and TP53 mutations as prognostic variables in tumours from breast cancer patients. Acta oncologica. 2008;47(4):600–7. doi: 10.1080/02841860802047411. [DOI] [PubMed] [Google Scholar]
- 29.Milsom MD, Schiedlmeier B, Bailey J, Kim MO, Li D, Jansen M, et al. Ectopic HOXB4 overcomes the inhibitory effect of tumor necrosis factor-{alpha} on Fanconi anemia hematopoietic stem and progenitor cells. Blood. 2009 May 21;113(21):5111–20. doi: 10.1182/blood-2008-09-180224. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Molecular cell. 2008 Dec 26;32(6):767–77. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Gurtan AM, Stuckert P, D’Andrea AD. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J Biol Chem. 2006 Apr 21;281(16):10896–905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- 32.Lensch MW, Tischkowitz M, Christianson TA, Reifsteck CA, Speckhart SA, Jakobs PM, et al. Acquired FANCA dysfunction and cytogenetic instability in adult acute myelogenous leukemia. Blood. 2003 Jul 1;102(1):7–16. doi: 10.1182/blood-2002-09-2781. [DOI] [PubMed] [Google Scholar]
- 33.Hess CJ, Ameziane N, Schuurhuis GJ, Errami A, Denkers F, Kaspers GJ, et al. Hypermethylation of the FANCC and FANCL promoter regions in sporadic acute leukaemia. Cellular oncology : the official journal of the International Society for Cellular Oncology. 2008;30(4):299–306. doi: 10.3233/CLO-2008-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009 Jul 2;114(1):174–80. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rio P, Segovia JC, Hanenberg H, Casado JA, Martinez J, Gottsche K, et al. In vitro phenotypic correction of hematopoietic progenitors from Fanconi anemia group A knockout mice. Blood. 2002 Sep 15;100(6):2032–9. [PubMed] [Google Scholar]
- 36.Li A, Yang Y, Gao C, Lu J, Jeong HW, Liu BH, et al. A SALL4/MLL/HOXA9 pathway in murine and human myeloid leukemogenesis. The Journal of clinical investigation. 2013 Oct 1;123(10):4195–207. doi: 10.1172/JCI62891. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000 Mar 9;404(6774):193–7. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 38.Will B, Siddiqi T, Jorda MA, Shimamura T, Luptakova K, Staber PB, et al. Apoptosis induced by JAK2 inhibition is mediated by Bim and enhanced by the BH3 mimetic ABT-737 in JAK2 mutant human erythroid cells. Blood. 2010 Apr 8;115(14):2901–9. doi: 10.1182/blood-2009-03-209544. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao C, Miyazaki M, Ohashi R, Tsuji T, Inoue Y, Namba M. Maintenance of near-diploid karyotype of PA-1 human ovarian teratocarcinoma cells due to death of polyploid cells by chromosome fragmentation/pulverization. International journal of molecular medicine. 1999 Sep;4(3):291–4. doi: 10.3892/ijmm.4.3.291. [DOI] [PubMed] [Google Scholar]
- 40.Rego MA, Kolling FWt, Vuono EA, Mauro M, Howlett NG. Regulation of the Fanconi anemia pathway by a CUE ubiquitin-binding domain in the FANCD2 protein. Blood. 2012 Sep 6;120(10):2109–17. doi: 10.1182/blood-2012-02-410472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huard CC, Tremblay CS, Magron A, Levesque G, Carreau M. The Fanconi anemia pathway has a dual function in Dickkopf-1 transcriptional repression. Proceedings of the National Academy of Sciences of the United States of America. 2014 Feb 11;111(6):2152–7. doi: 10.1073/pnas.1314226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui W, Kong NR, Ma Y, Amin HM, Lai R, Chai L. Differential expression of the novel oncogene, SALL4, in lymphoma, plasma cell myeloma, and acute lymphoblastic leukemia. Mod Pathol. 2006 Dec;19(12):1585–92. doi: 10.1038/modpathol.3800694. eng. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.