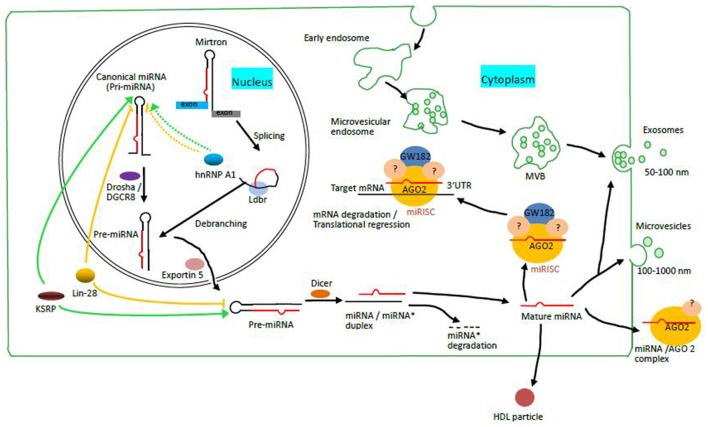
Figure 1.
MiRNA biosynthesis through canonical and non-canonical pathways and the mechanisms of miRNA release into the bloodstream (Modified from Zandberga et al., 2013). Mirtrons are spliced, and then debranched by lariat debranching enzyme (Ldbr) to generate pre-miRNAs, the product of Drosha/DGCR 8 cleavage of pri-miRNAs in canonical pathways. Pre-miRNAs from both pathways are exported into the cytoplasm by Exportin 5 for further processing by Dicer, thus producing double-stranded miRNA/miRNA* duplexes. Subsequently, one strand of this duplex is loaded into miRNA induced silencing complex (miRISC) containing one of four AGOs (such as AGO2 as shown), GW182 and various unknown GW182-interacting silencing effectors, leaving the other strand undergoing degradation. The mature miRNA then guides the miRISC to bind to the 3′ UTR of target mRNAs either to promote mRNA degradation or to inhibit protein translation. KSRP as a co-activator and Lin-28 as a co-repressor bind to the terminal loop (TL) elements of pri or pre-miRNAs to promote and inhibit the maturation of a subset of miRNA precursors respectively. HnRNP A1 can both promote (for pri-miR-18a) and inhibit (for pri-let-7a) the processing of miRNAs by binding to the TL elements of pri-miRNAs as shown in the figure, which is mediated by different mechanisms. Circulating miRNAs exist either as a vesicle-associated form or as a protein-associated form in the circulation. The former (vesicles) include exosomes and microvesicles. The latter (proteins) include high and low density lipoproteins, RNA binding proteins such as Argonaute 2.