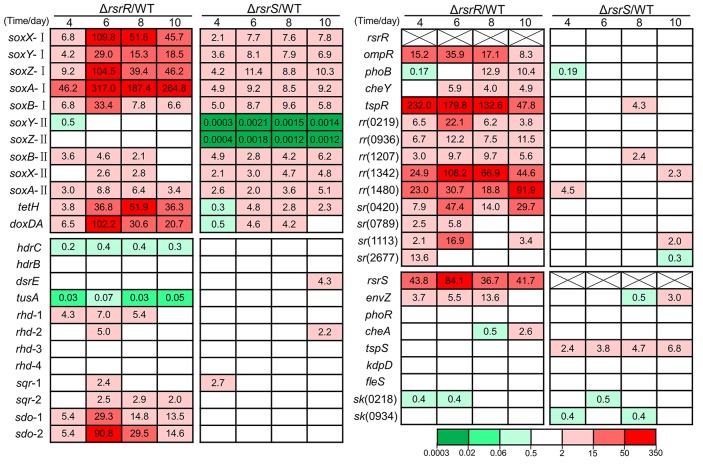
Figure 7.
The relative transcription levels of genes involved in sulfur metabolism and signaling systems during the S0-cultivating process. This is the valid mean value of three independent replicates showing fold changes (FC) determined by RT-qPCR analyses of the mutant against the wild-type. FC ≥ 2, P ≤ 0.05 and FC ≤ 0.5, P ≤ 0.05 were regarded as the significant changes. FC ≥ 2, P ≤ 0.05, up-regulation; FC ≤ 0.5, P ≤ 0.05, down-regulation; 0.5 ≤ FC ≤ 2, P ≥ 0.05, no change (data are not shown in the figure). FC-values are represented with different colors as indicated in the color bar. The data of standard deviation (SD) and P-value were shown in Supplementary Text S1. The SD-value was calculated by using the Origin software with “descriptive statistics.” The P-value was calculated by using the GraphPad Prism software with “unpaired t-test.” The original data with the values for standard deviation and P-value are listed in Supplementary Text S1. The putative function of proteins encoded by these genes: soxX-I (A5904_2486), soxX-II (A5904_2525), cytochrome c class I; soxY-I (A5904_2487), soxY-II (A5904_2520), sulfur covalently binding protein; soxZ-I (A5904_2488), soxZ-II (A5904_2521), sulfur compound chelating protein; soxA-I (A5904_2489), soxA-II (A5904_2526), cytochrome c (diheme); soxB-I (A5904_2491), soxB-II (A5904_2522), sulfate thiol esterase; hdrC (A5904_1042), hdrB (A5904_1043), heterodisulfide reductase subunit C and B; dsrE (A5904_2473), tusA (A5904_2474), sulfur transferase; rhd-1 (A5904_0894), rhd-2 (A5904_1407), rhd-3 (A5904_2860), rhd-4 (A5904_2475), rhodanese (sulfur transferase); sqr-1 (A5904_1436), sqr-2 (A5904_2678), sulfide quinone reductase; sdo-1 (A5904_0421), sdo-2 (A5904_0790), sulfur dioxygenase; envZ (A5904_2589), ompR (A5904_2590), osmolarity regulation; phoB (A5904_0374), phoR (A5904_0373), phosphate regulon; cheY (A5904_1450), cheA (A5904_1448), chemotaxis; tspR (A5904_2485), tspS (A5904_2484), regulation for Sox pathway; kdpD (A5904_1340), unknown; fleS (A5904_1479), flagellum associated; rr (A5904_0219, A5904_0936, A5904_1207, A5904_1342, A5904_1480), putative response regulators in TCSs; hk (A5904_0218, A5904_0934), putative sensor histidine kinases in TCSs; sr (A5904_0420, A5904_0789, A5904_1113, A5904_2677), putative regulators in SCSs.