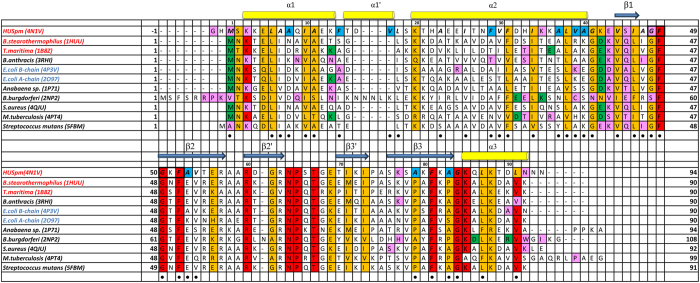
Figure 2. Multiple sequence alignment of HUSpm and HU proteins with known three-dimensional structures.
The PDB code is given in parentheses after the name of the organism. The name of the organism is red if its HU protein has high thermal stability and blue if its HU protein has low thermal stability. The HUSpm secondary structure elements are indicated. The residues involved in the formation of the DS region are in black frames. Residues involved in the formation of the hydrophobic core of HUSpm are marked with a black circle. Non-homologous residues of the HUSpm hydrophobic core are in blue. Residues that form hydrogen bonds in the dimers of HU proteins are in magenta and residues that form salt bridges in the dimers of HU proteins are in green. The N-terminal Met residue in all HU proteins, except for HUSpm and the HU protein from B. burgdorferi, forms both a salt bridge and a hydrogen bond.